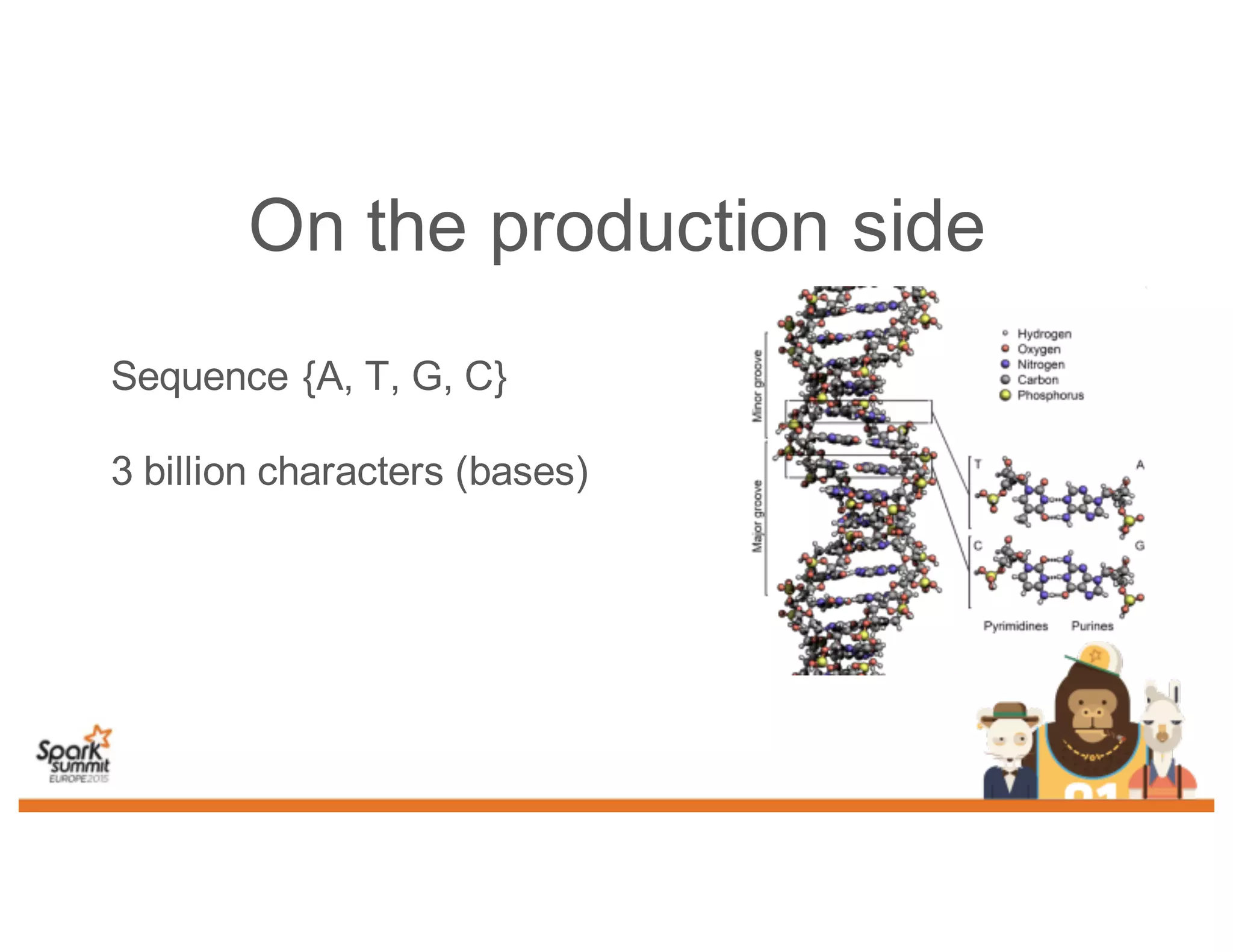
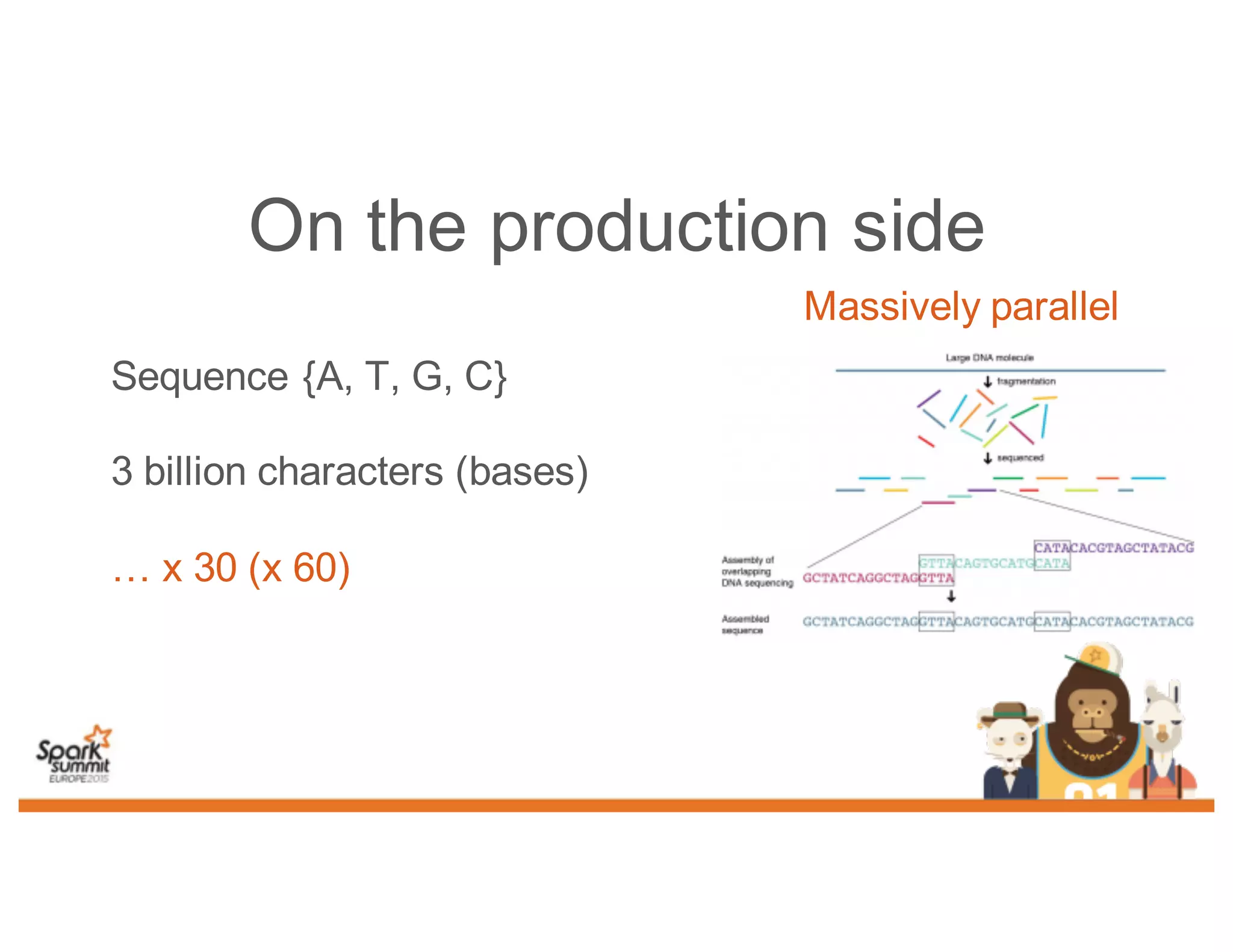
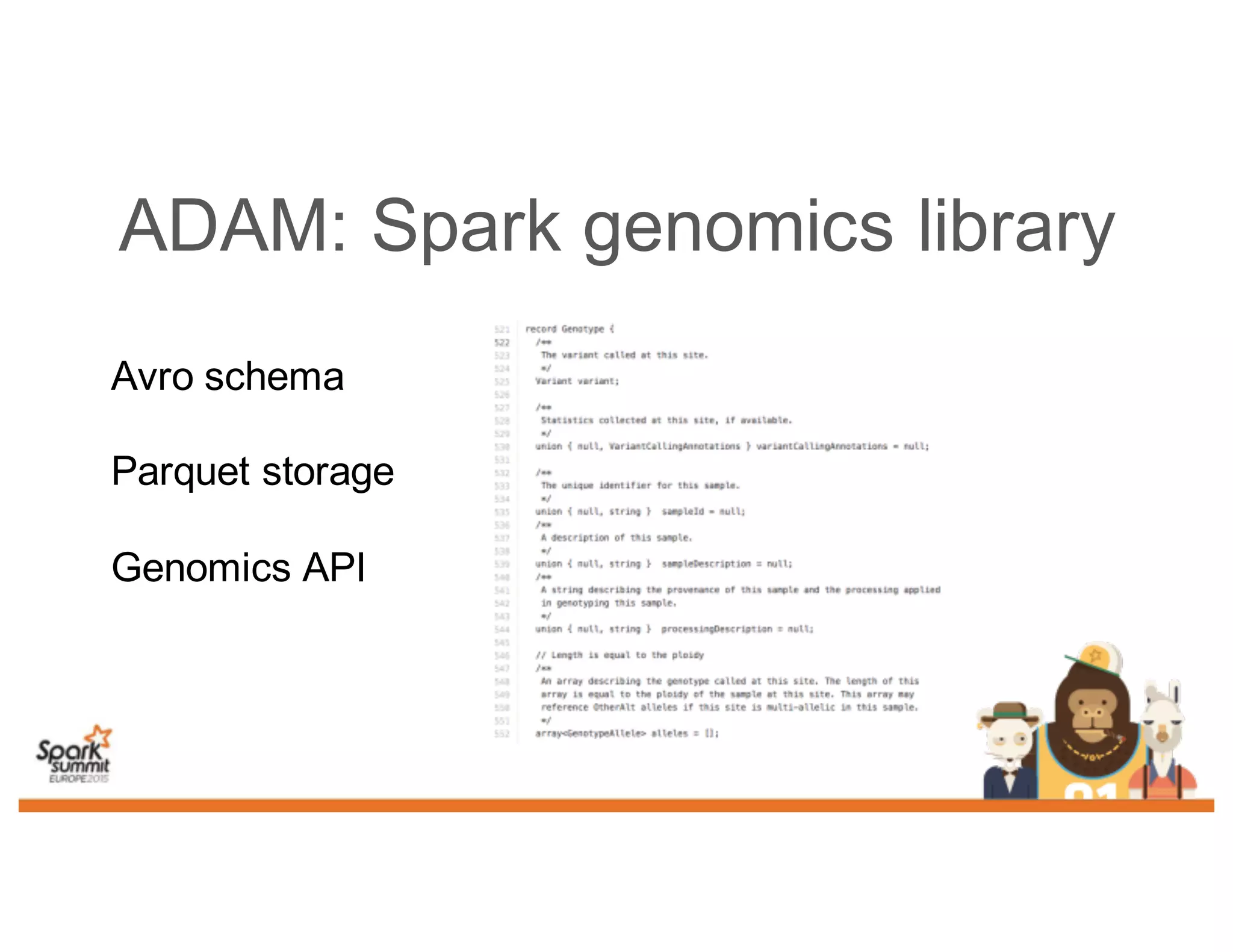
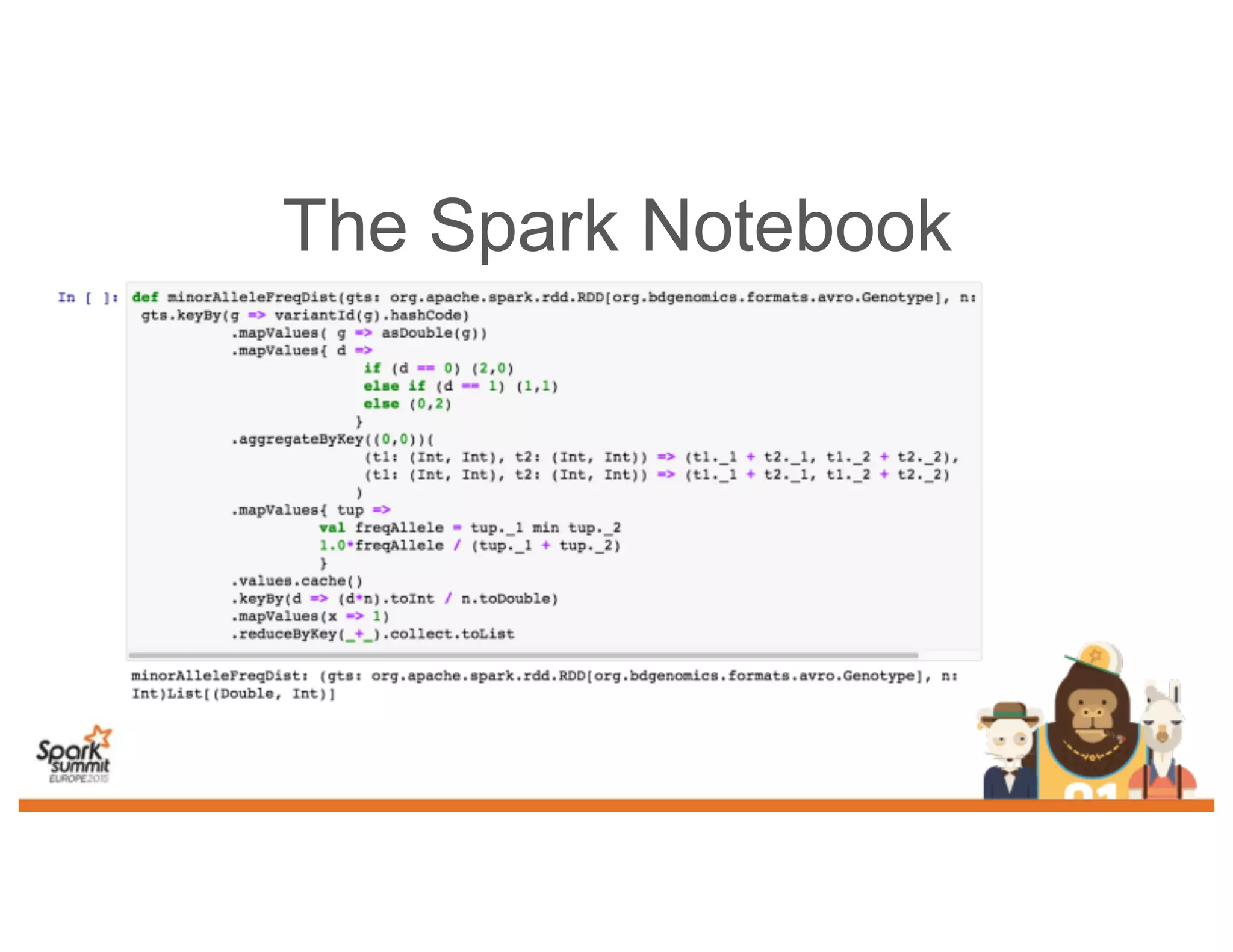
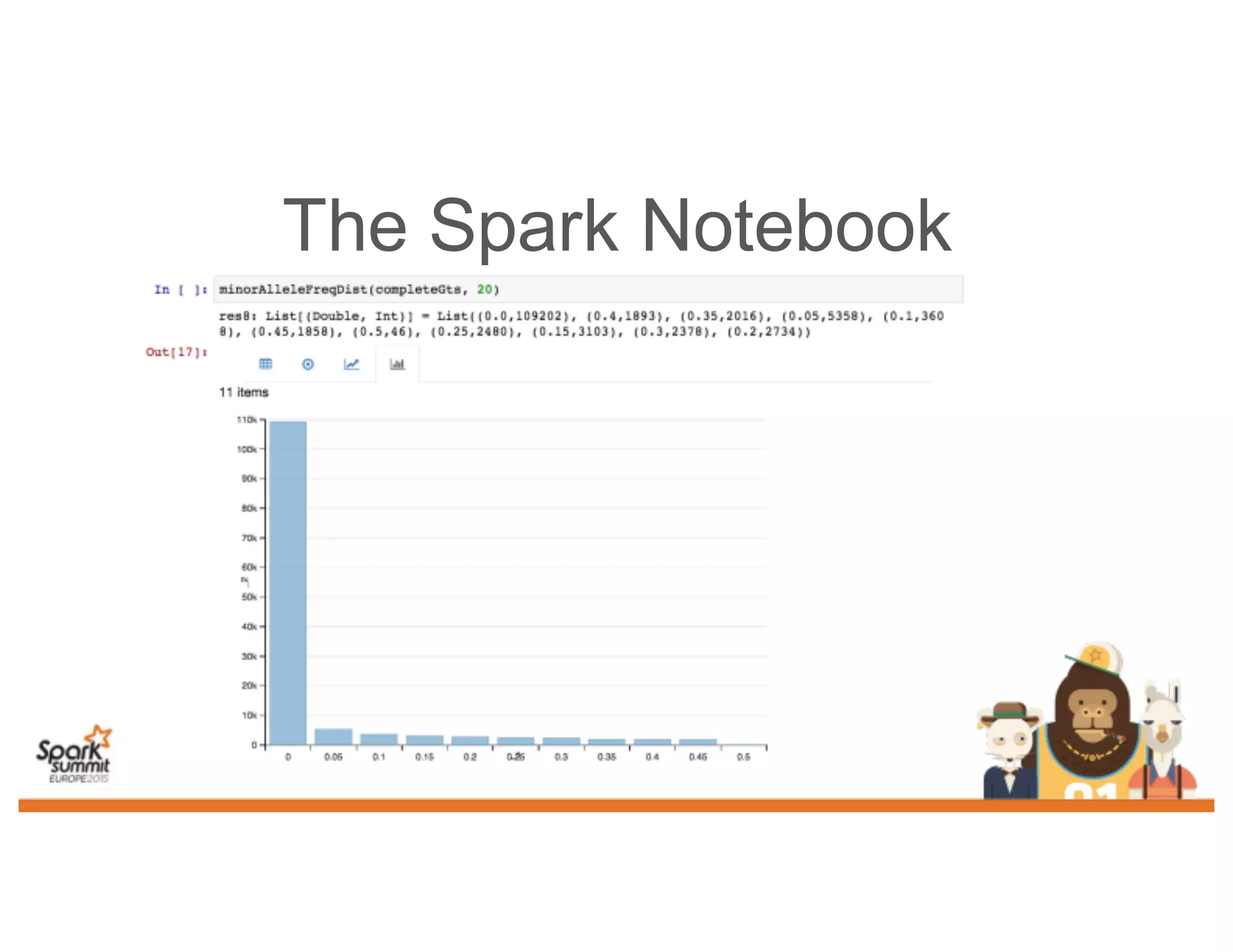
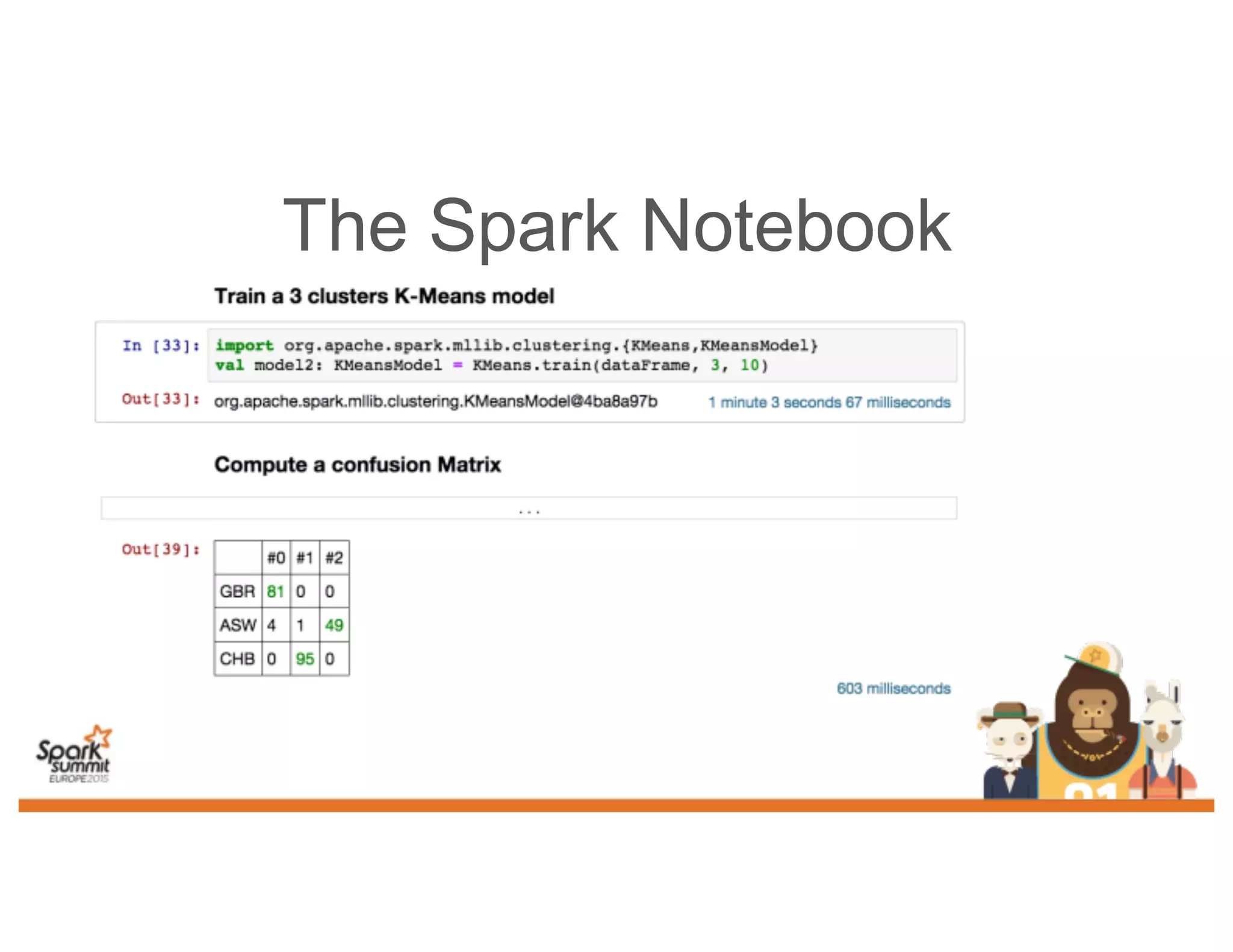
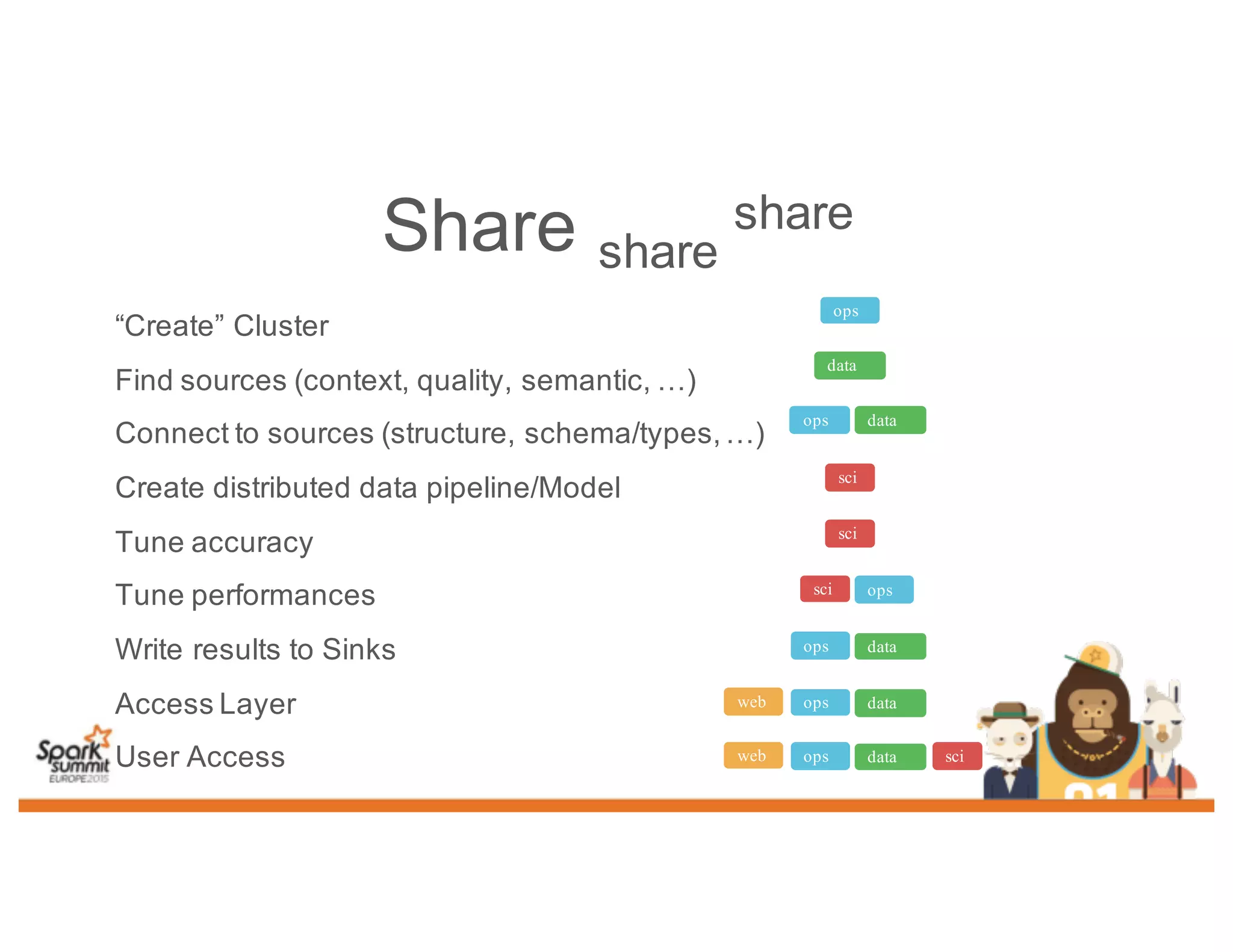
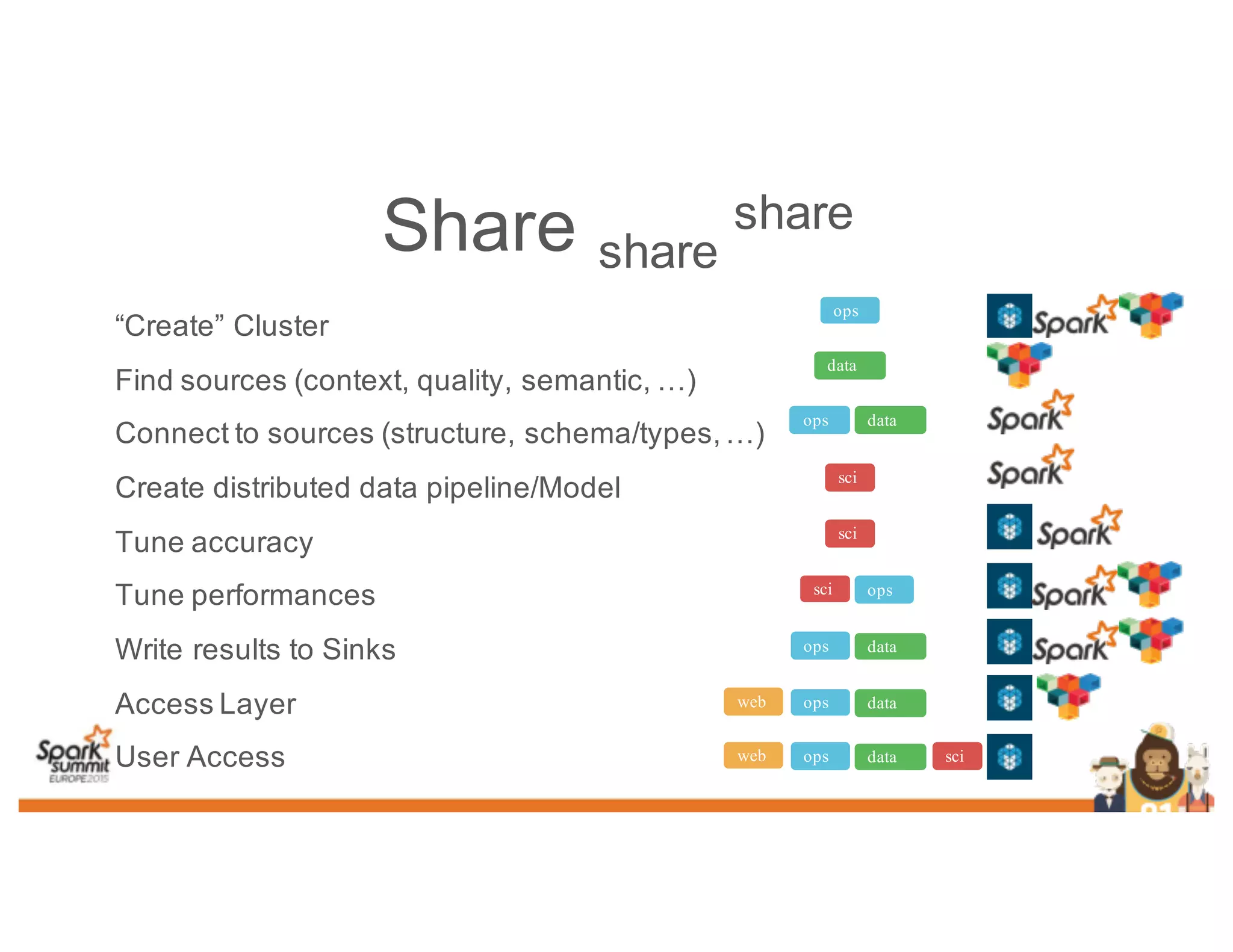
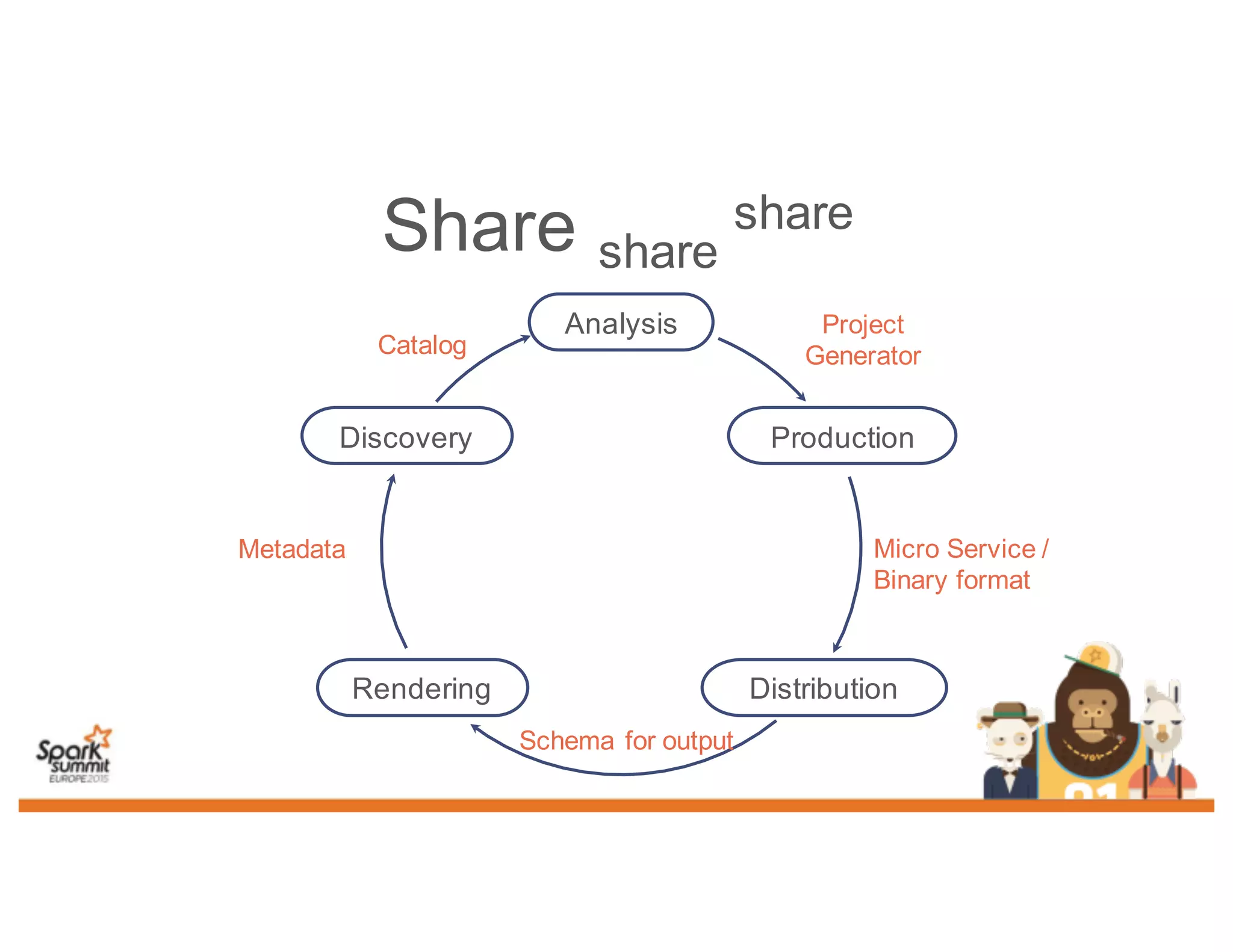
This document discusses analyzing genomic data at scale using distributed machine learning tools like Spark, ADAM, and the Spark Notebook. It outlines challenges with genomic data like its large size and need for distributed teams in research projects. The document proposes sharing data, processes, and results more efficiently through tools like Shar3 that can streamline the data analysis lifecycle and allow distributed collaboration on genomic research projects and datasets.