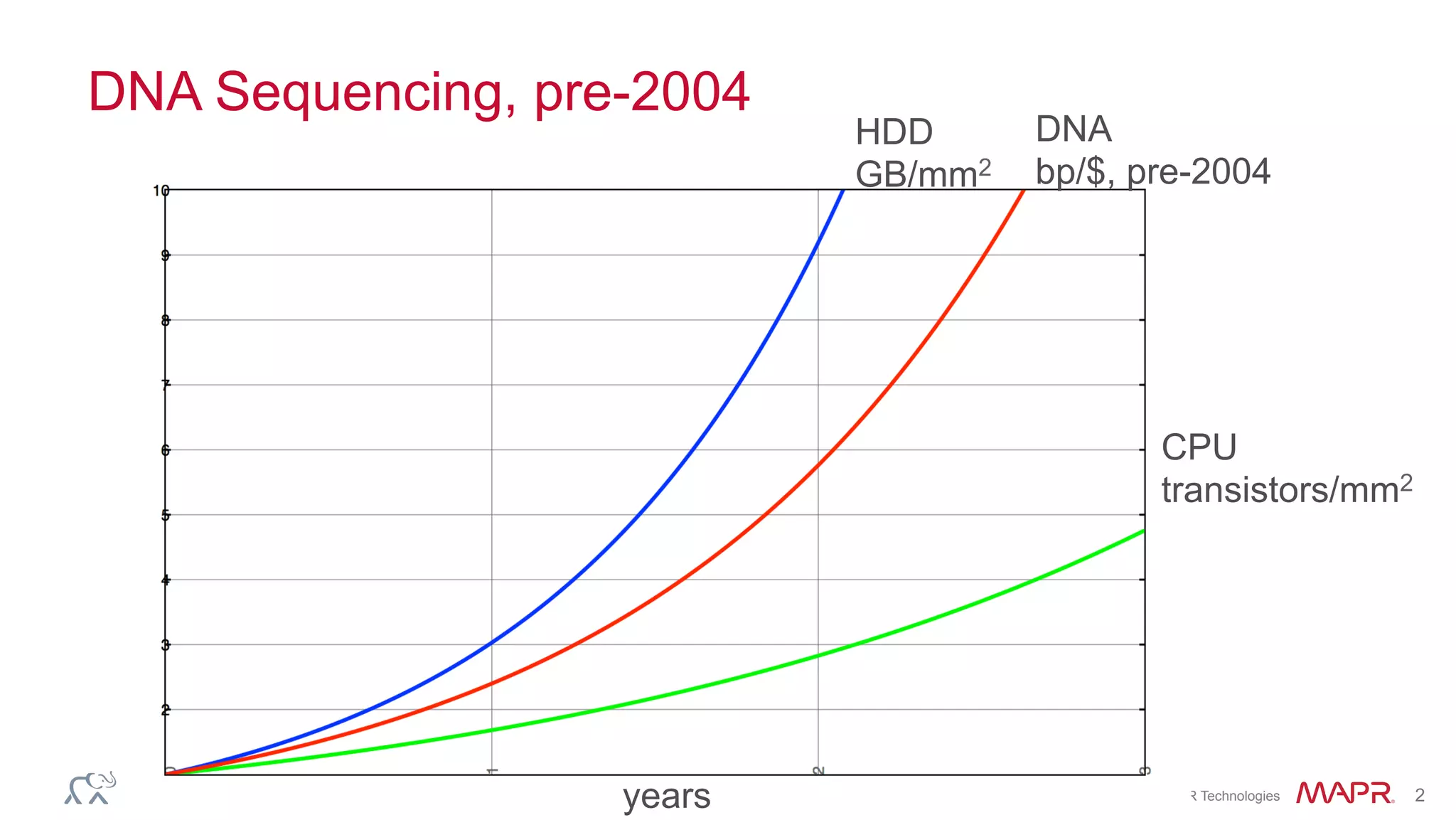
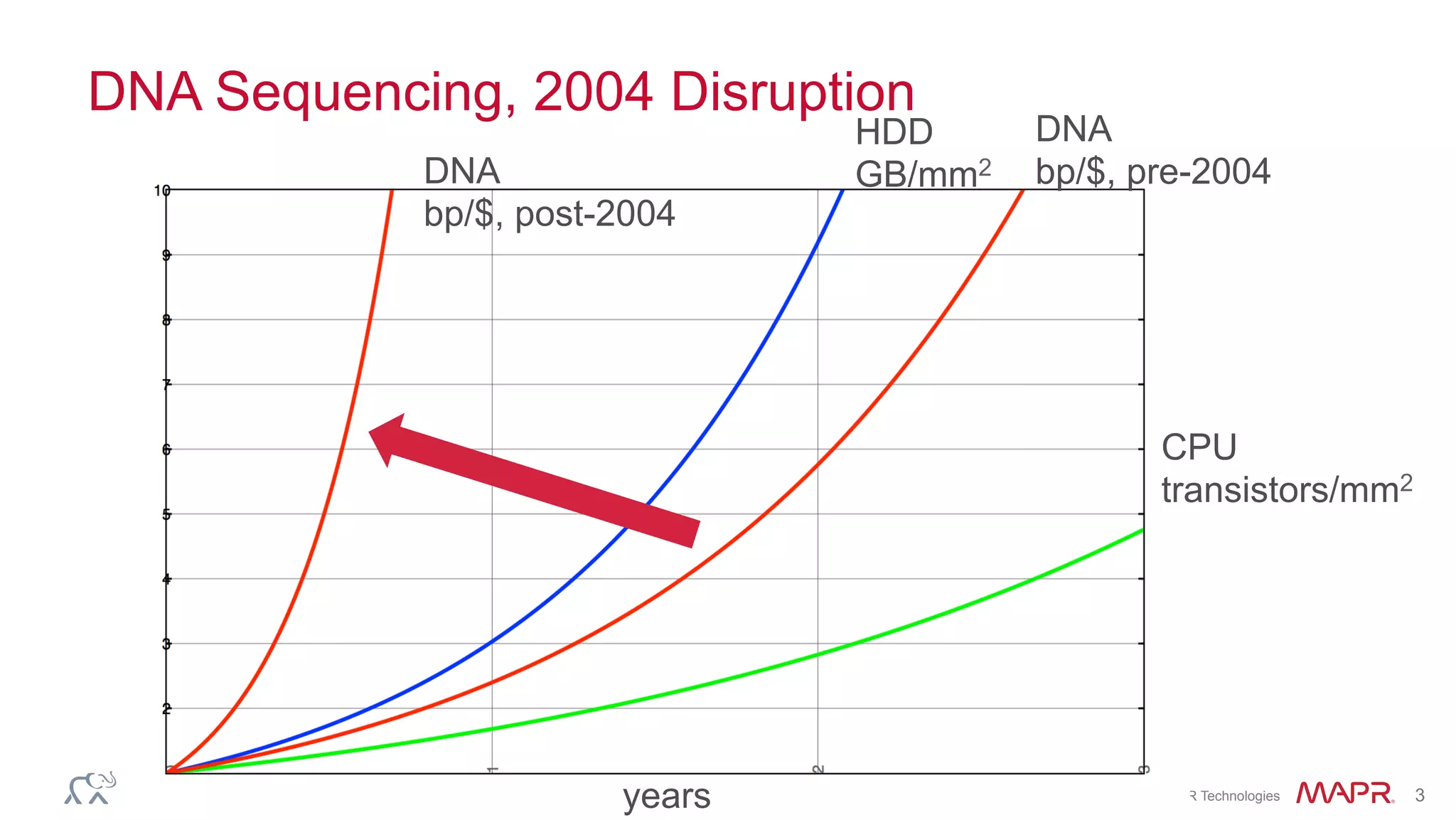
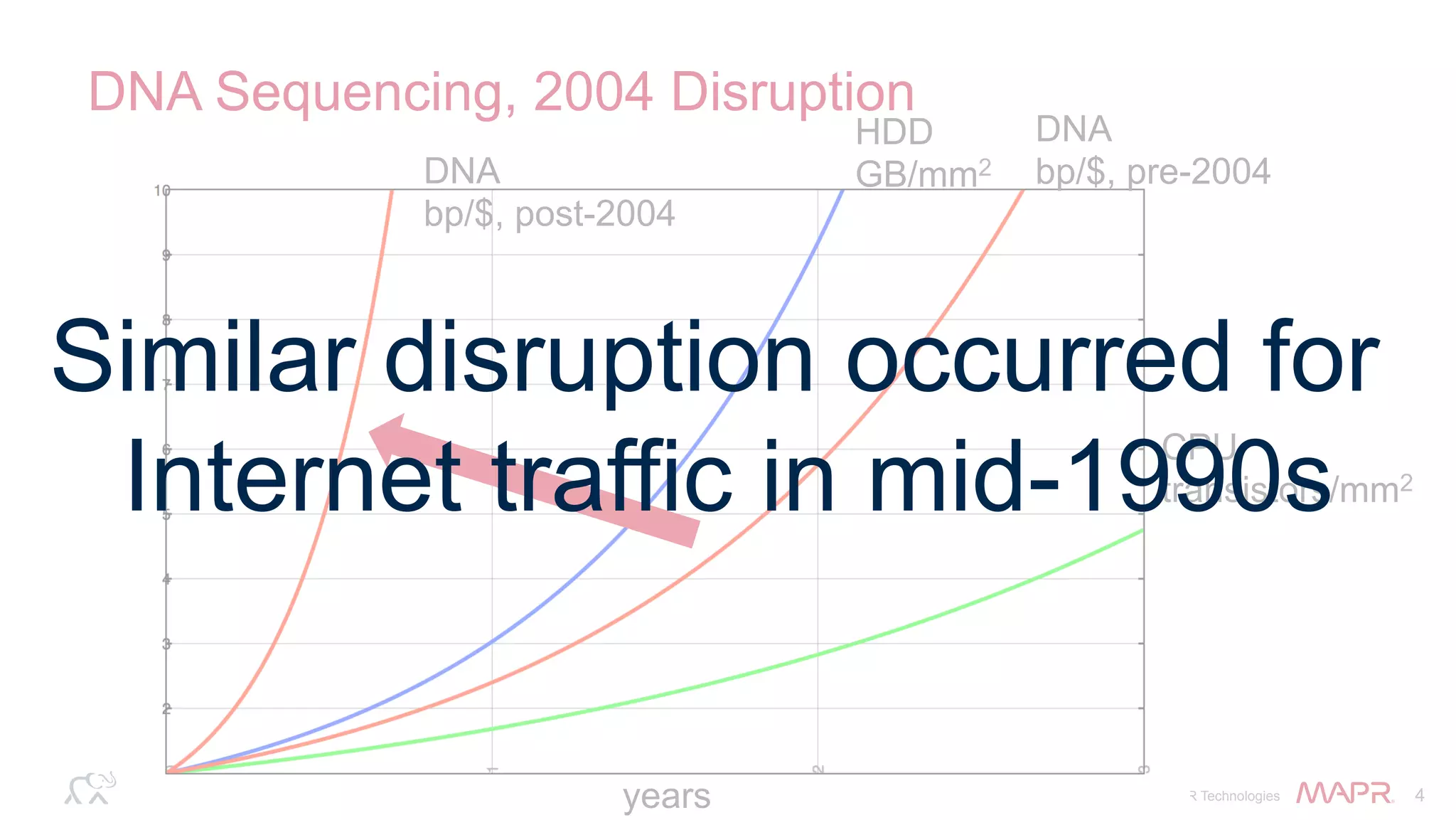
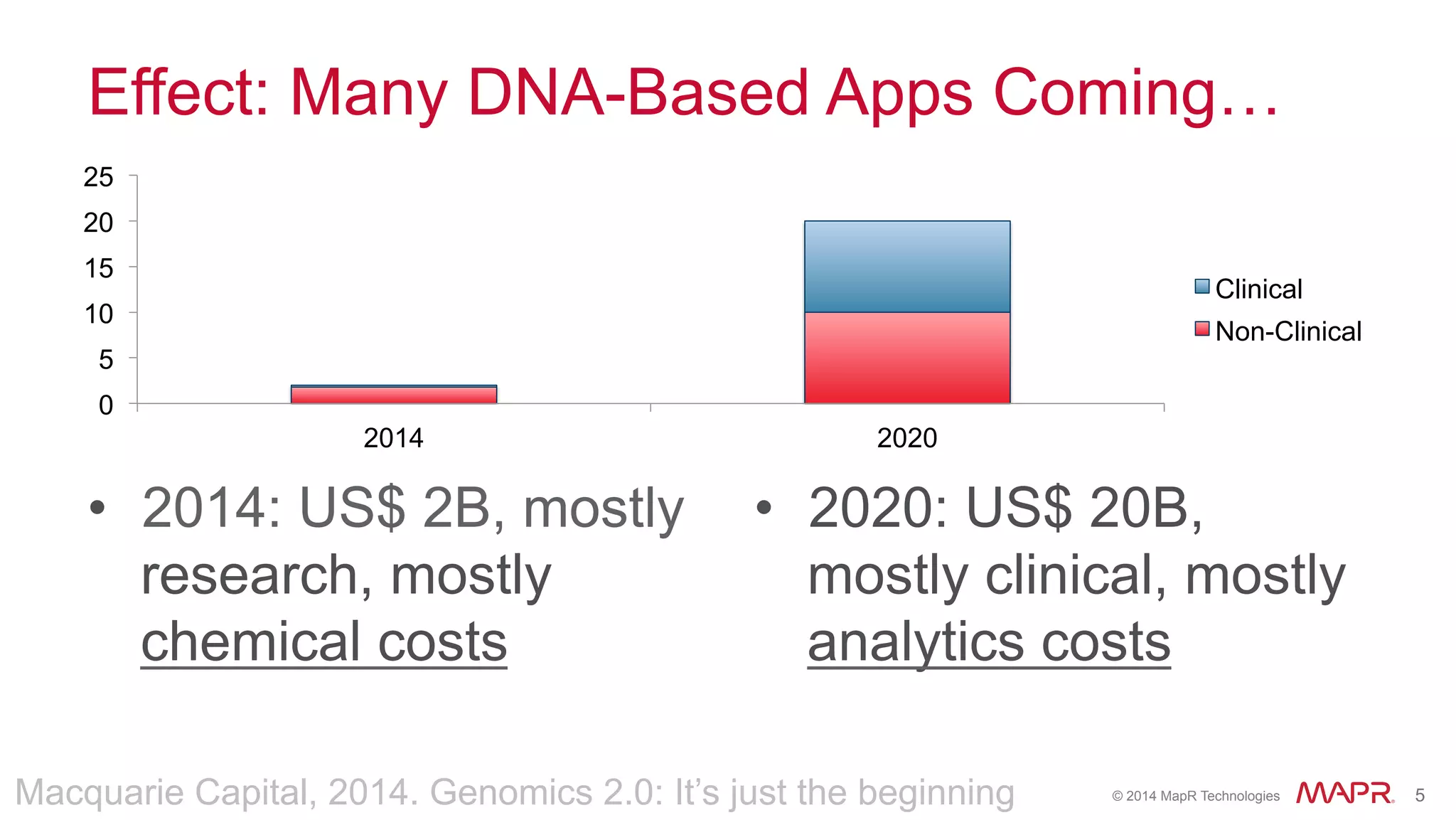
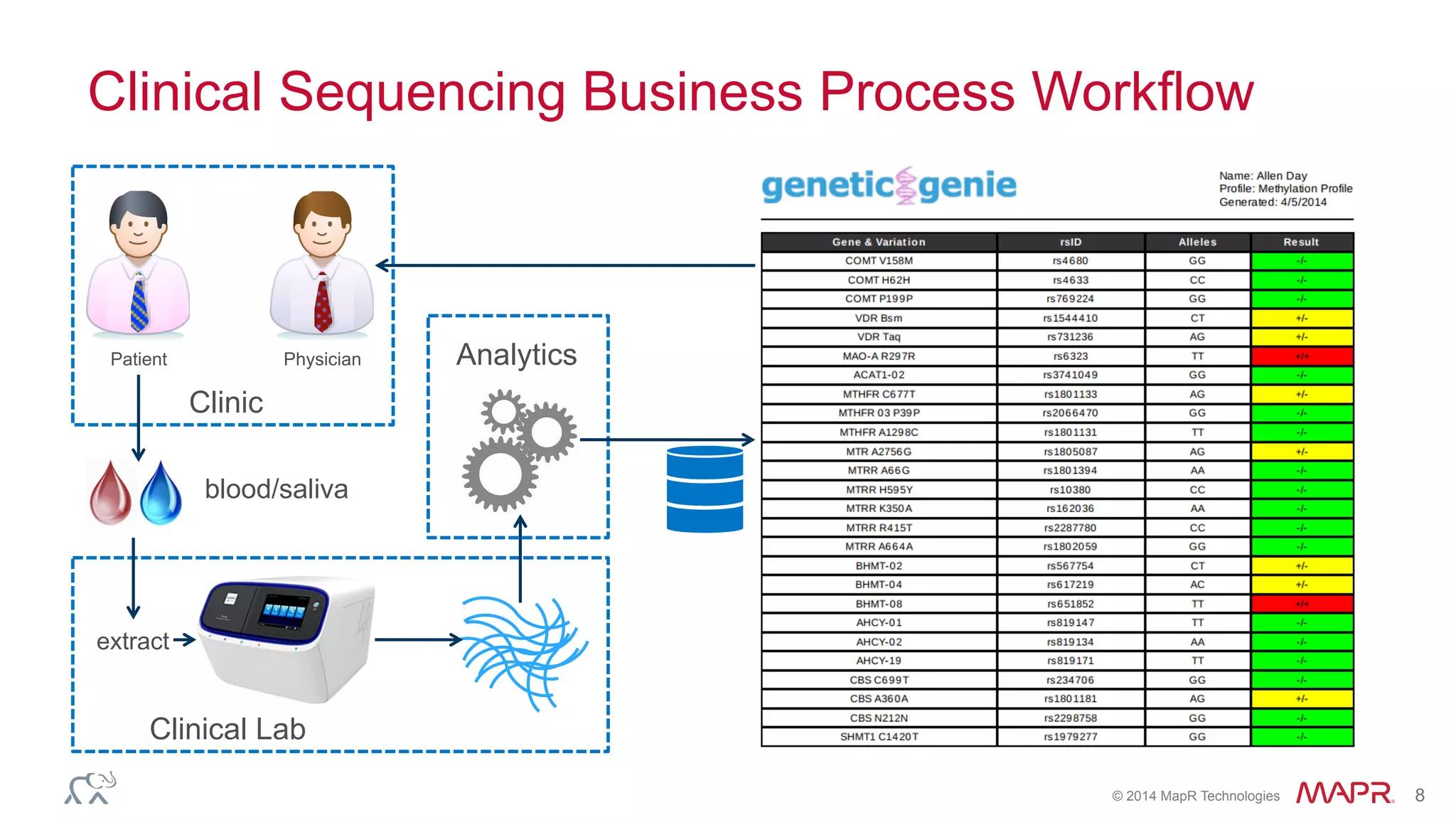
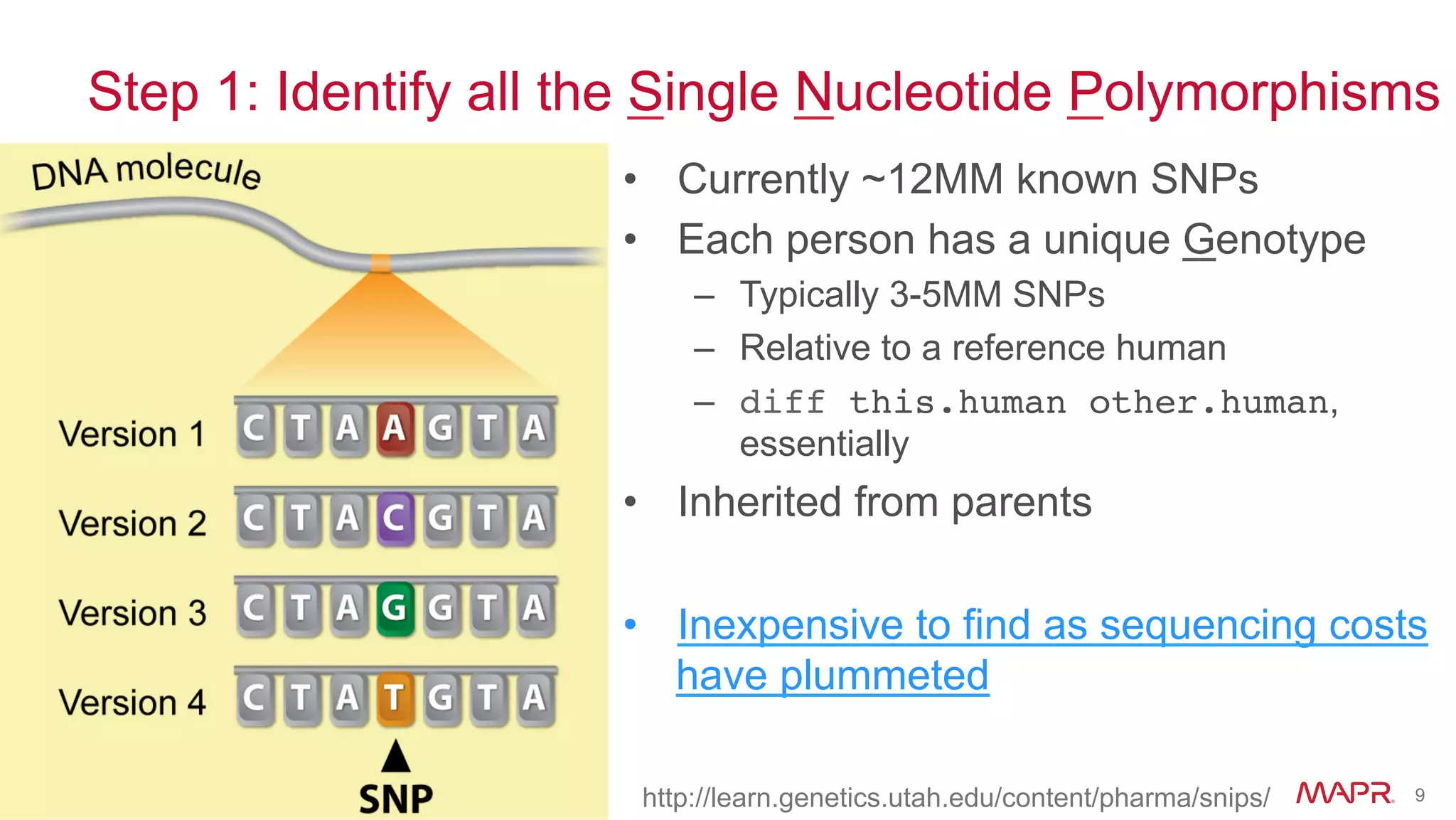
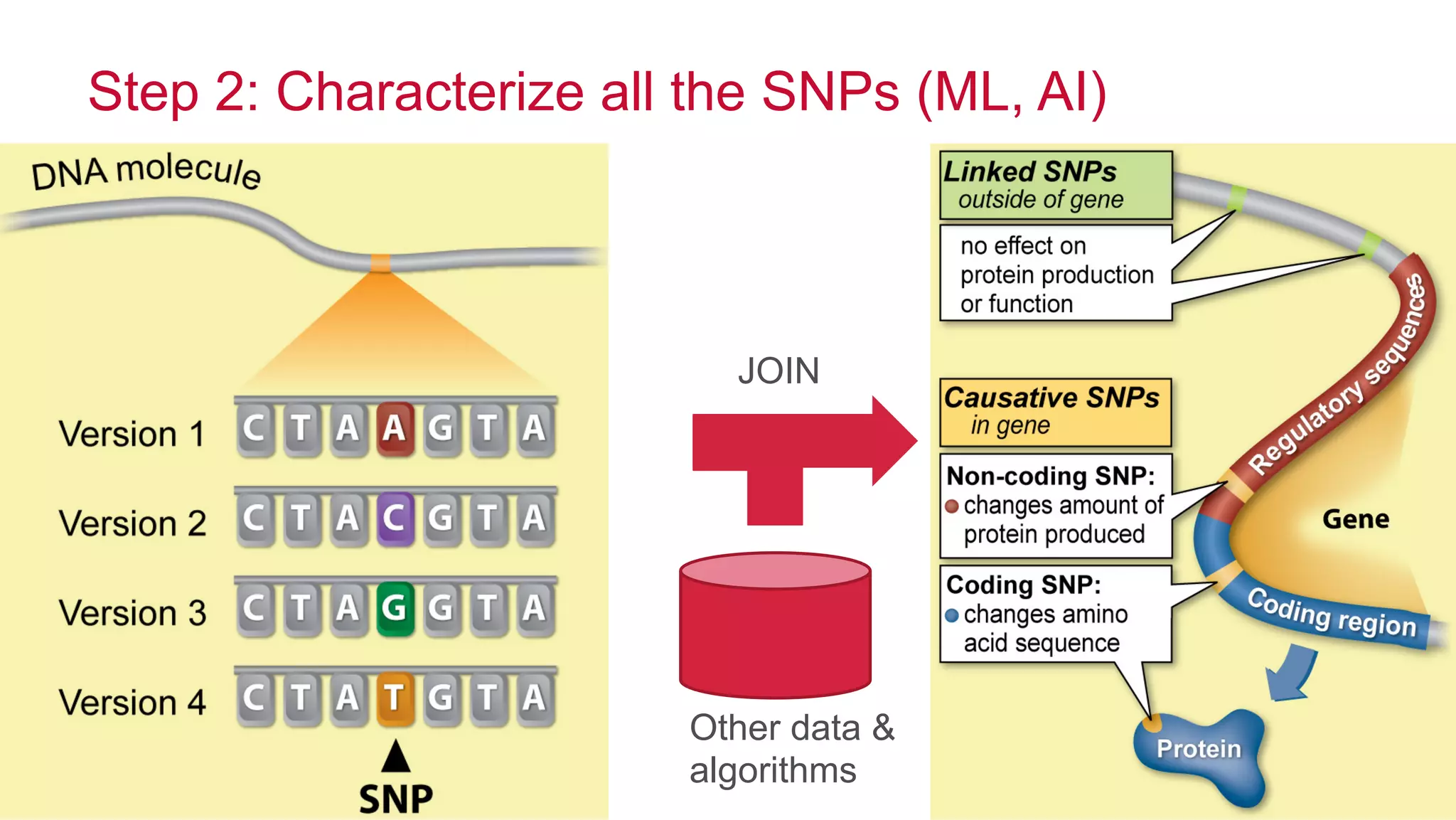
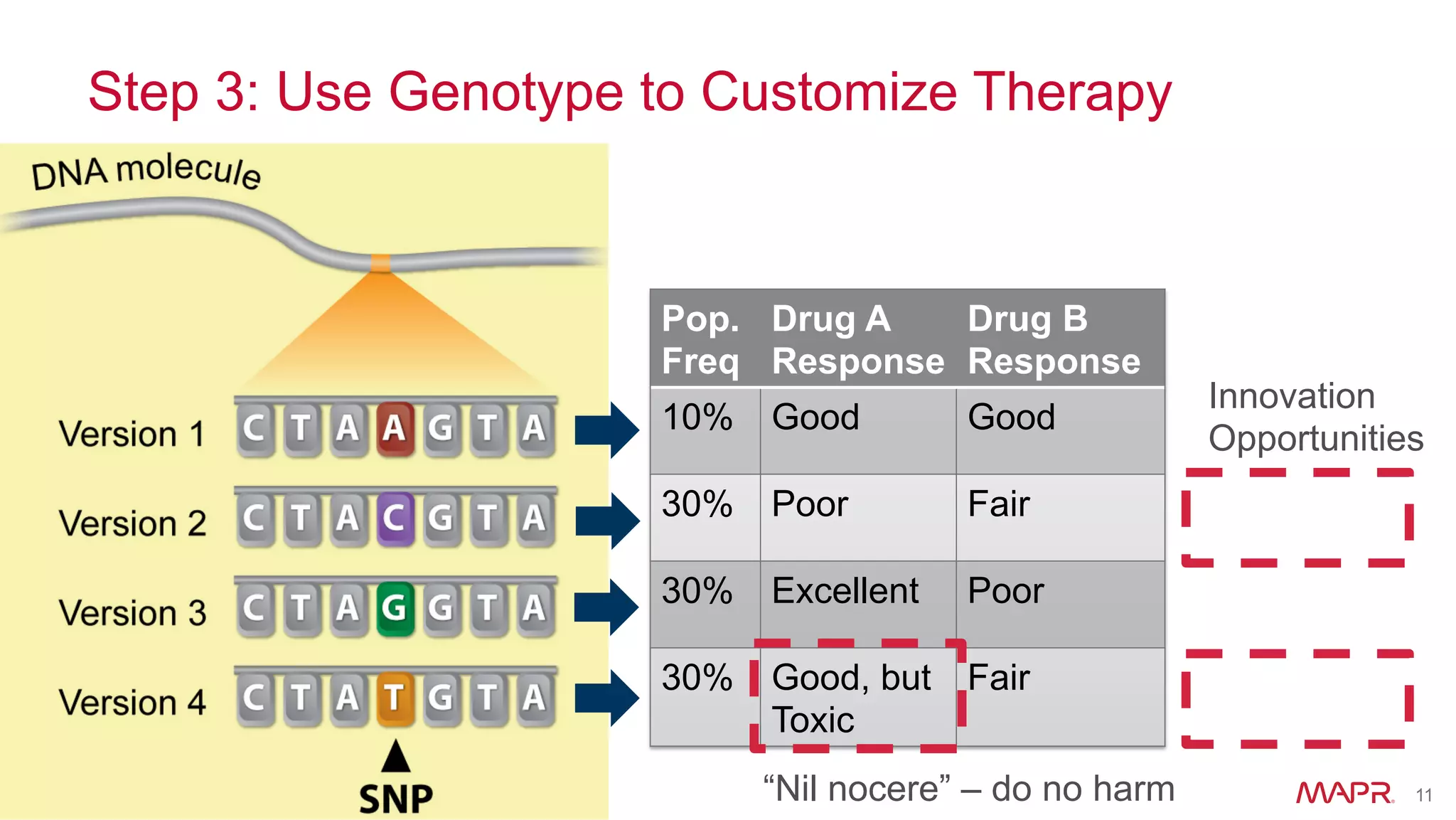
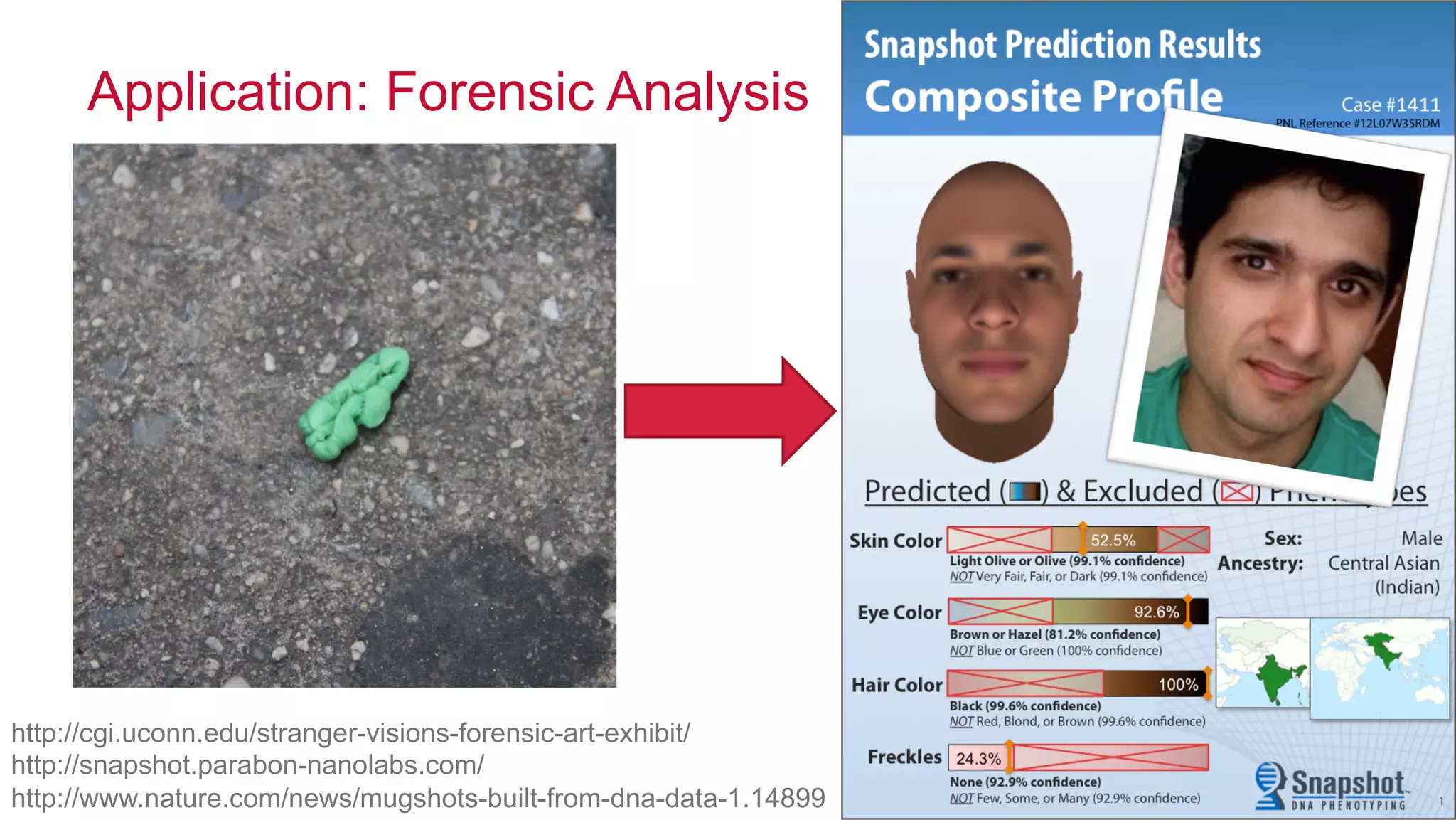
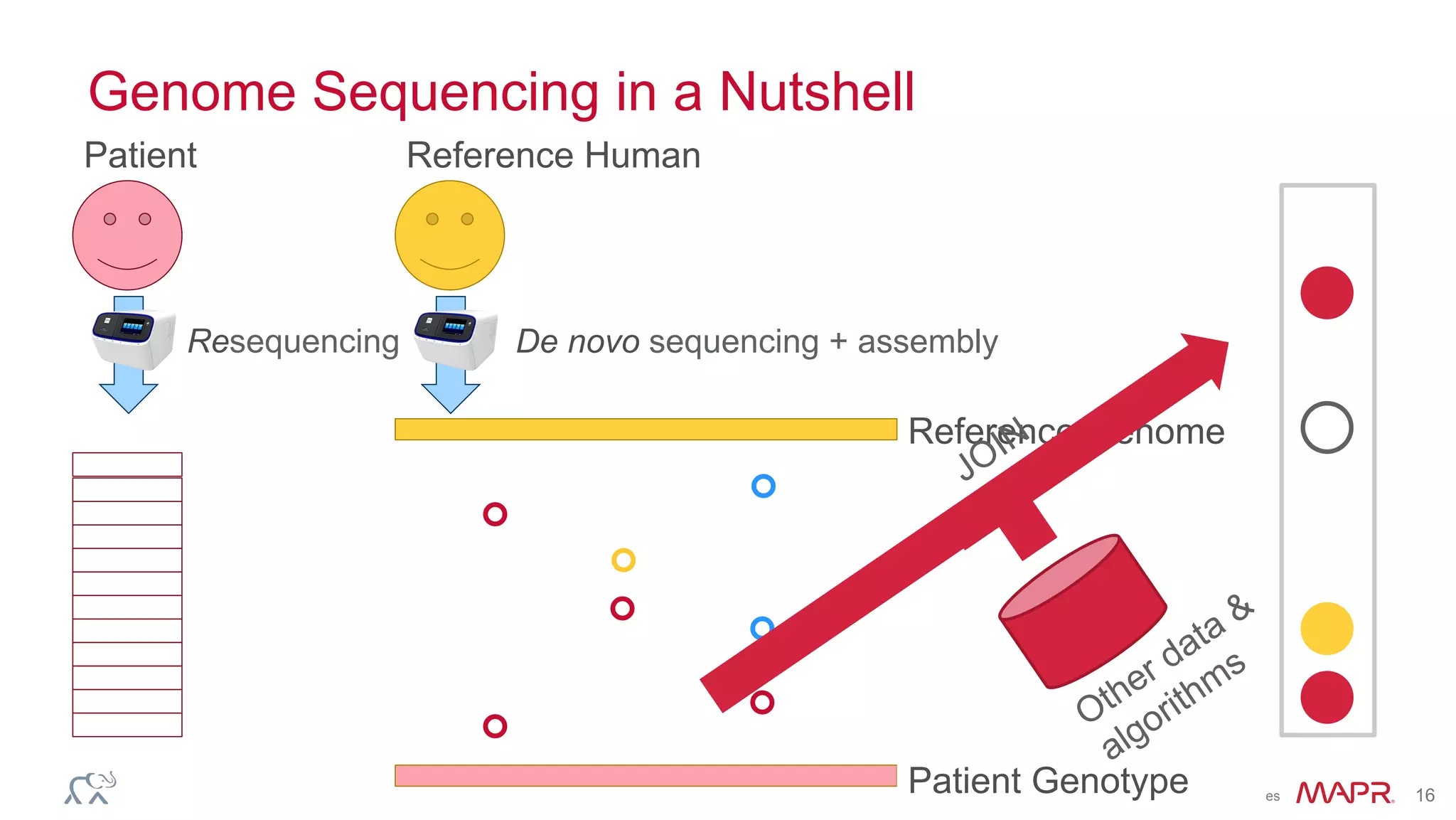
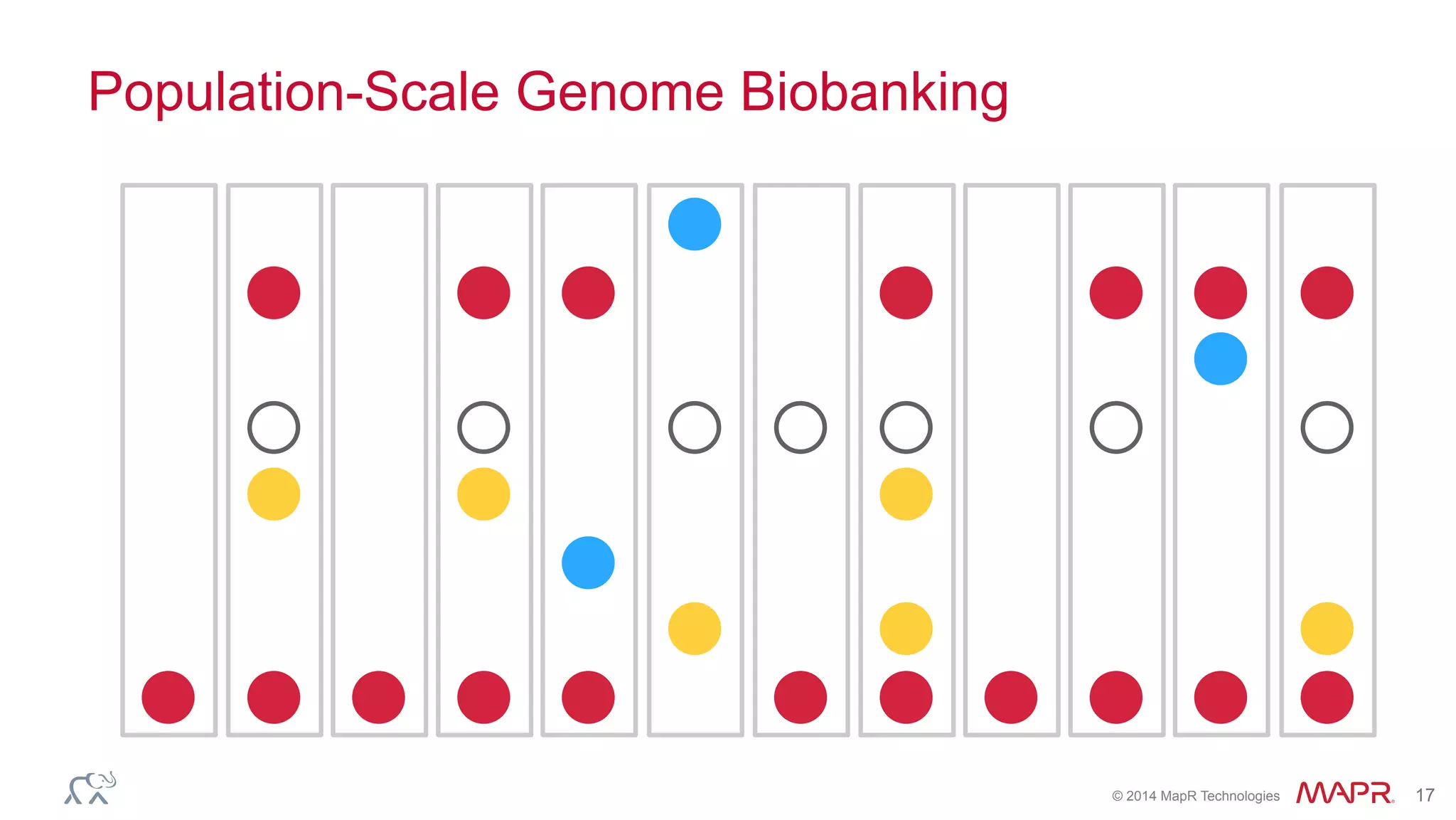
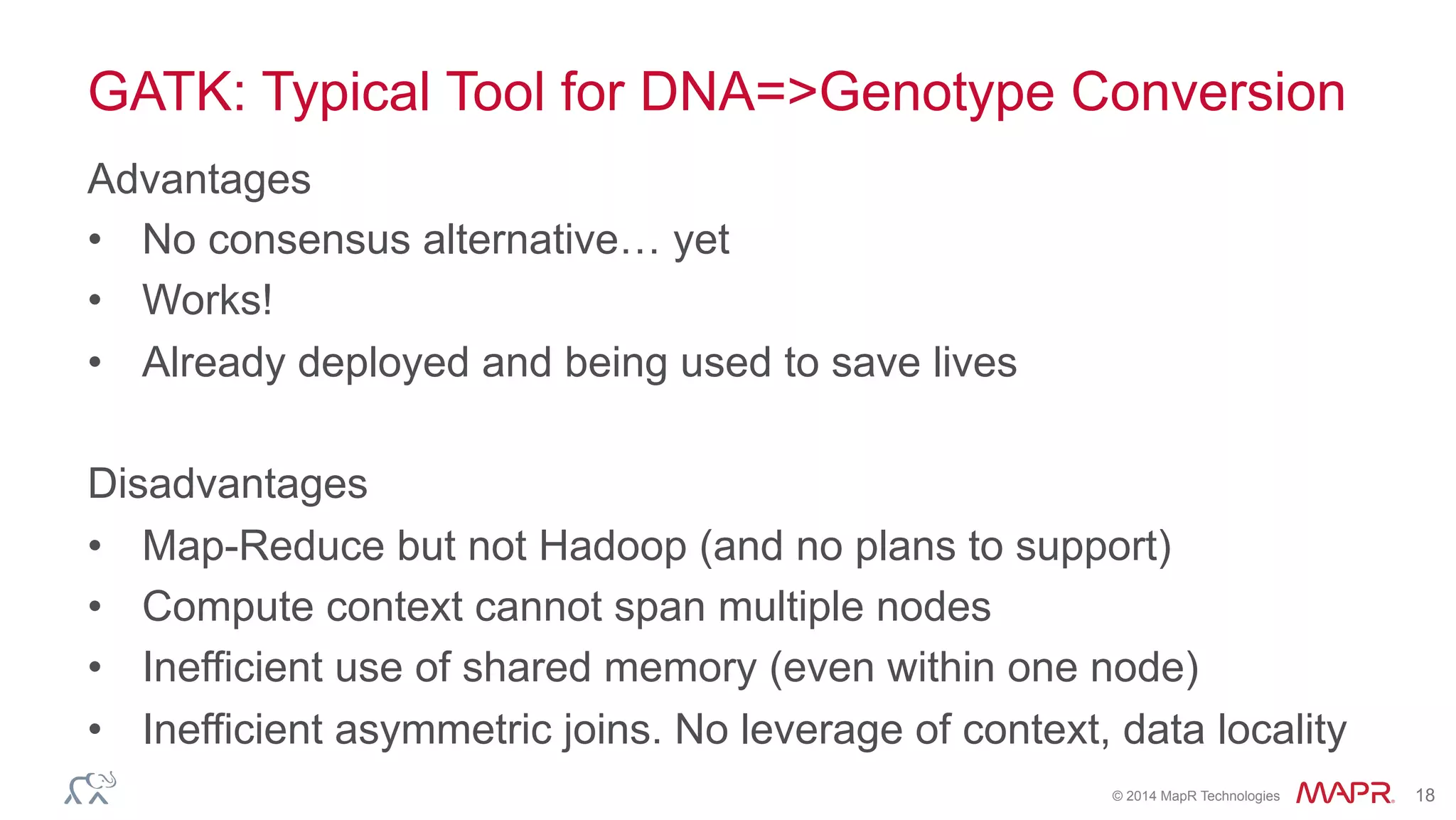
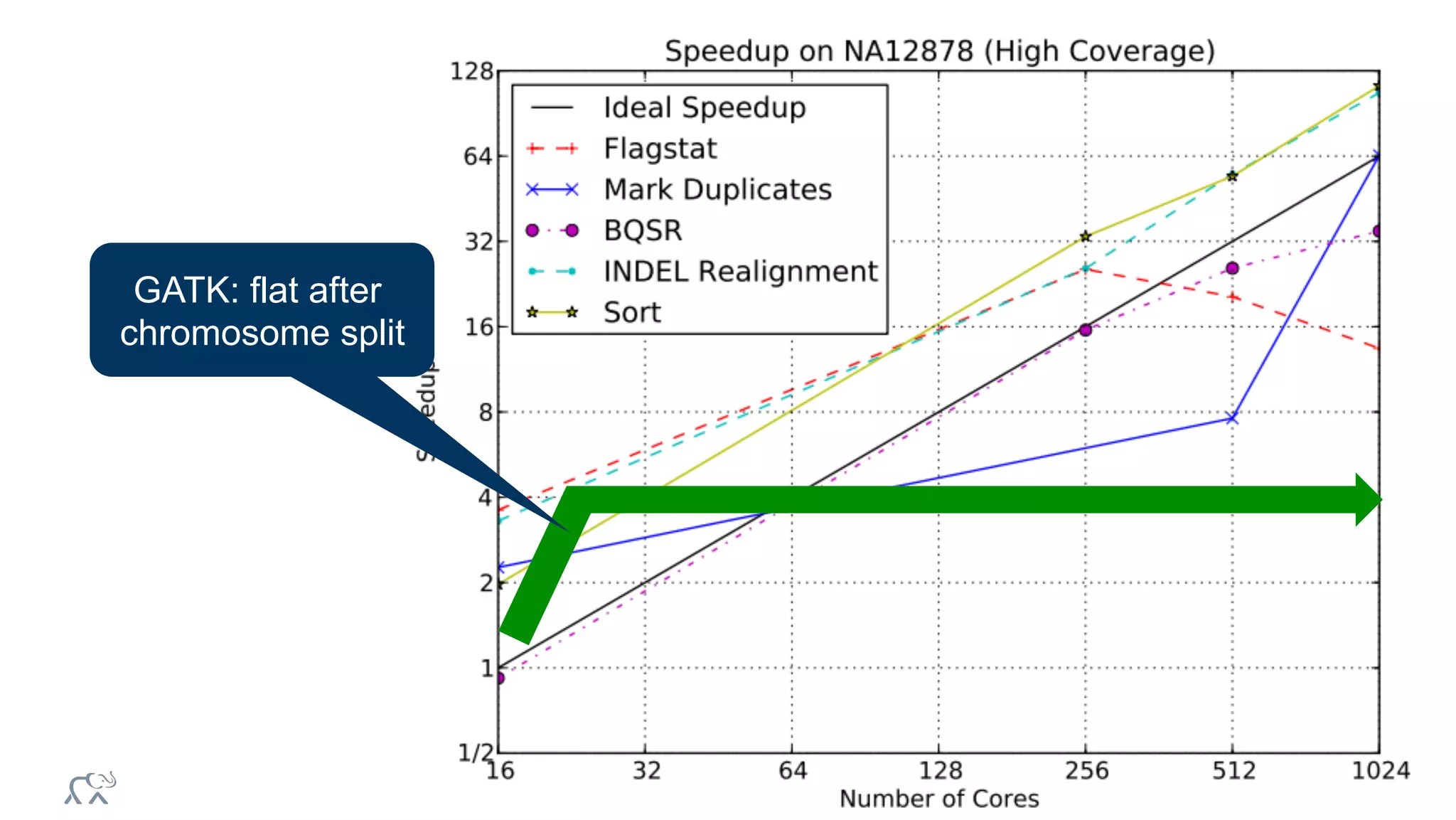
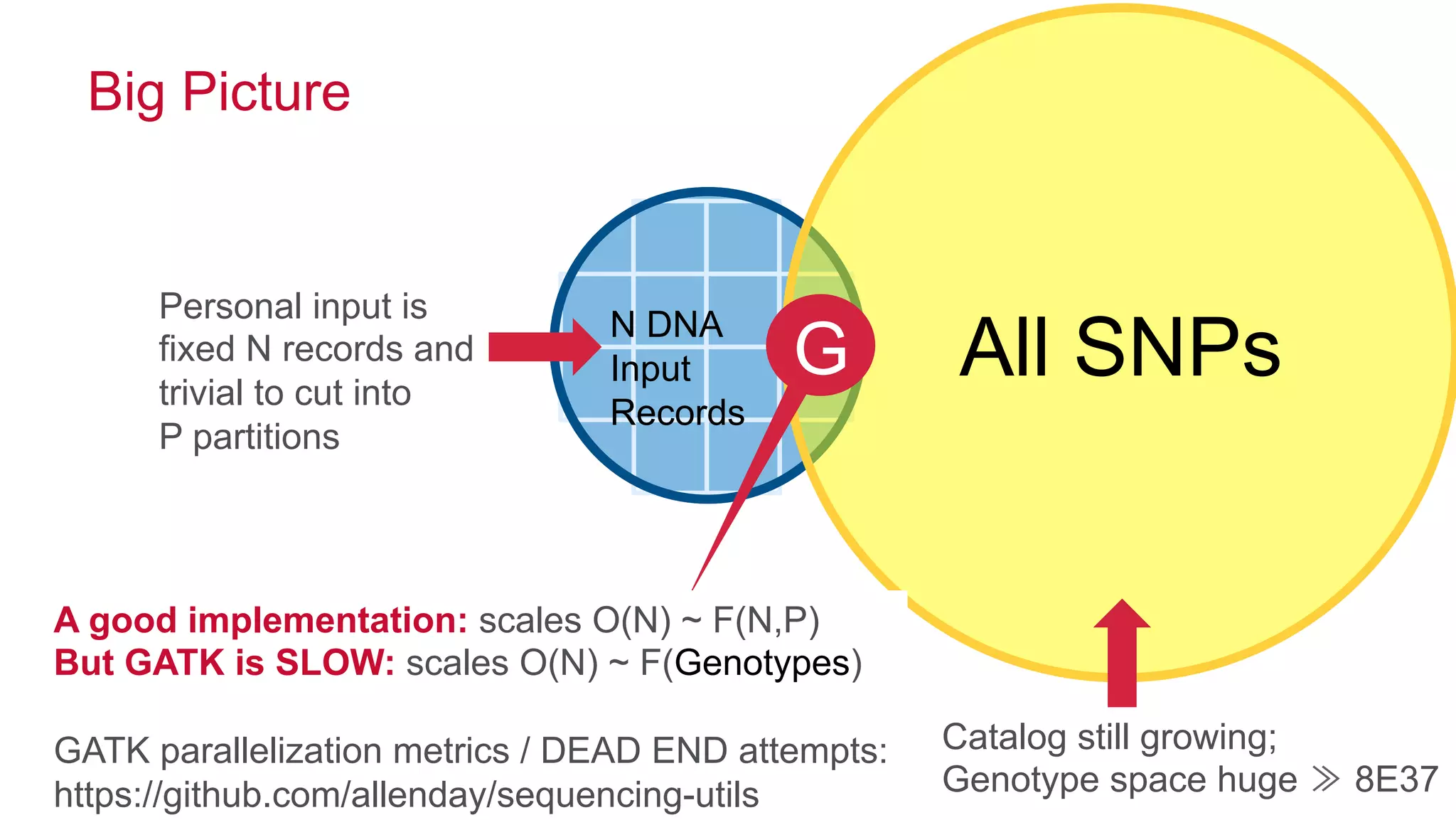
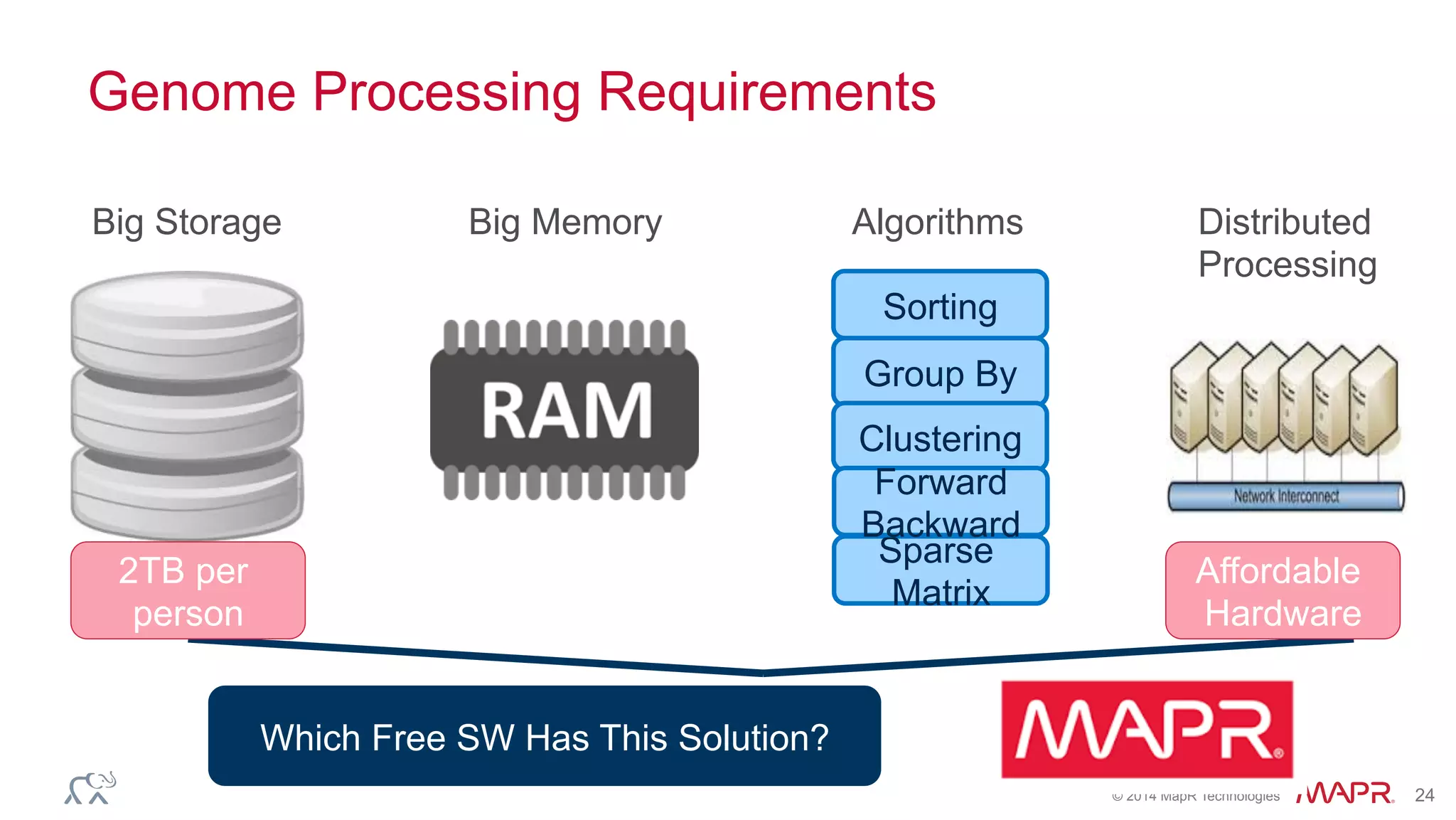
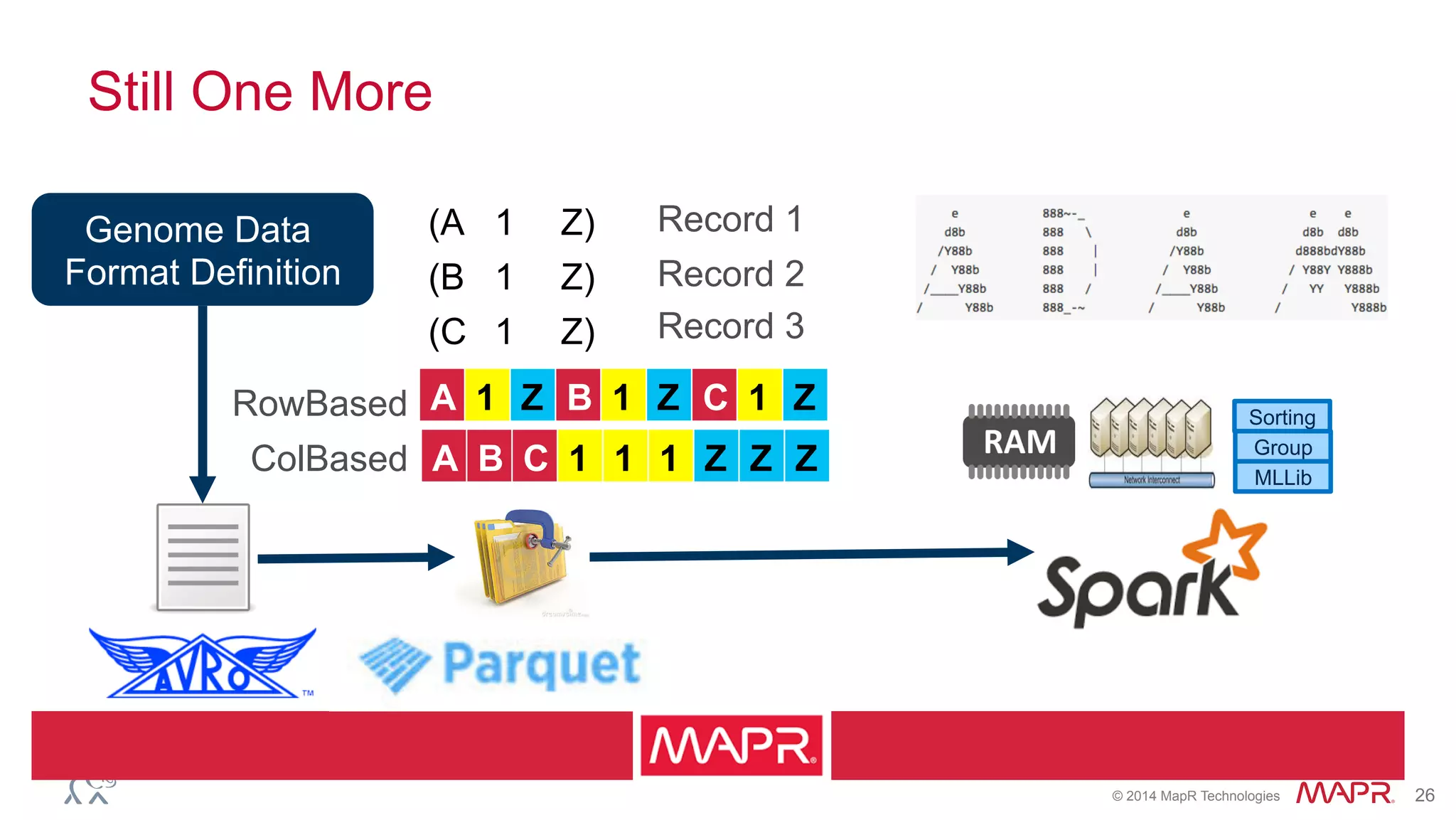
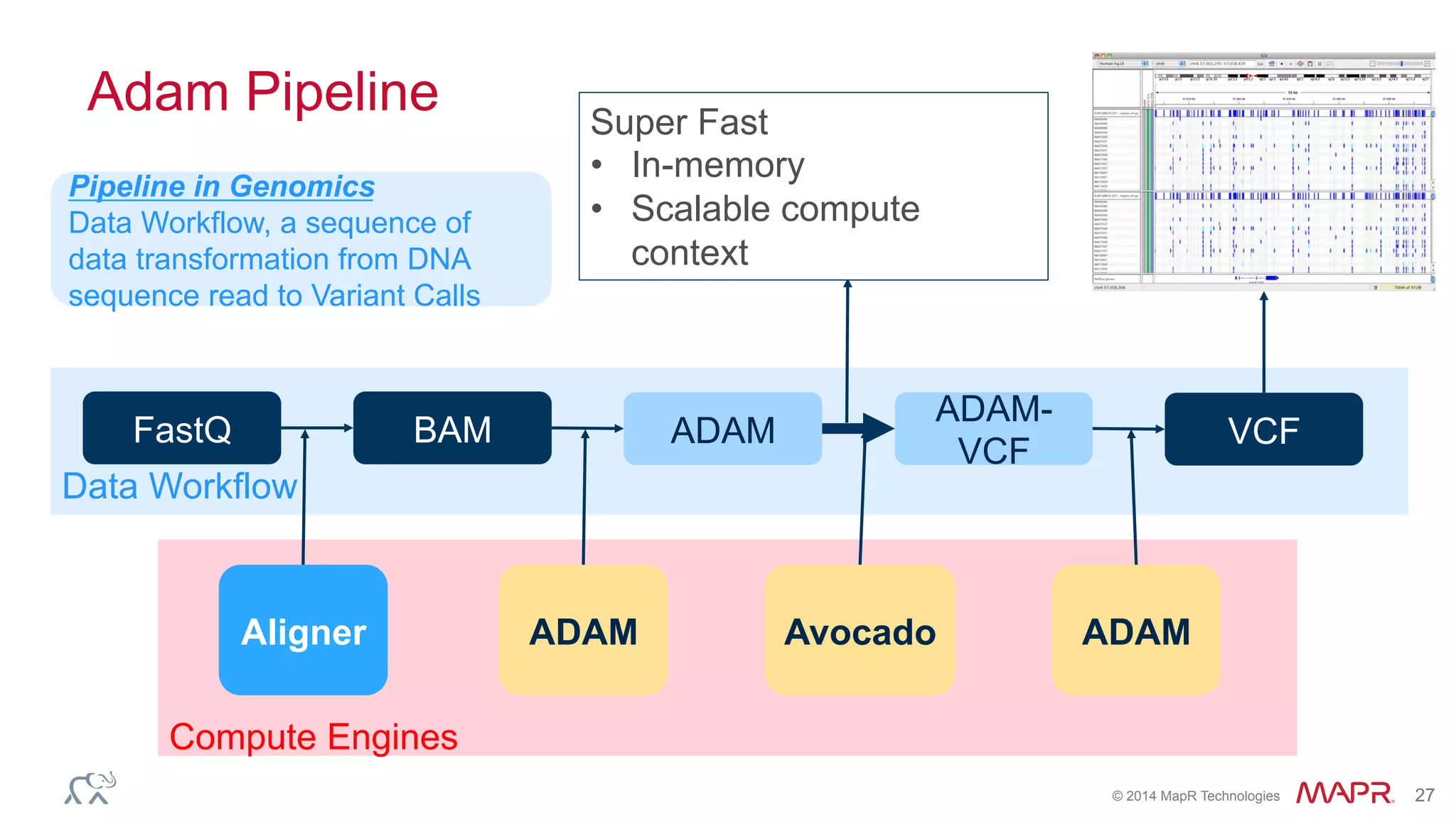
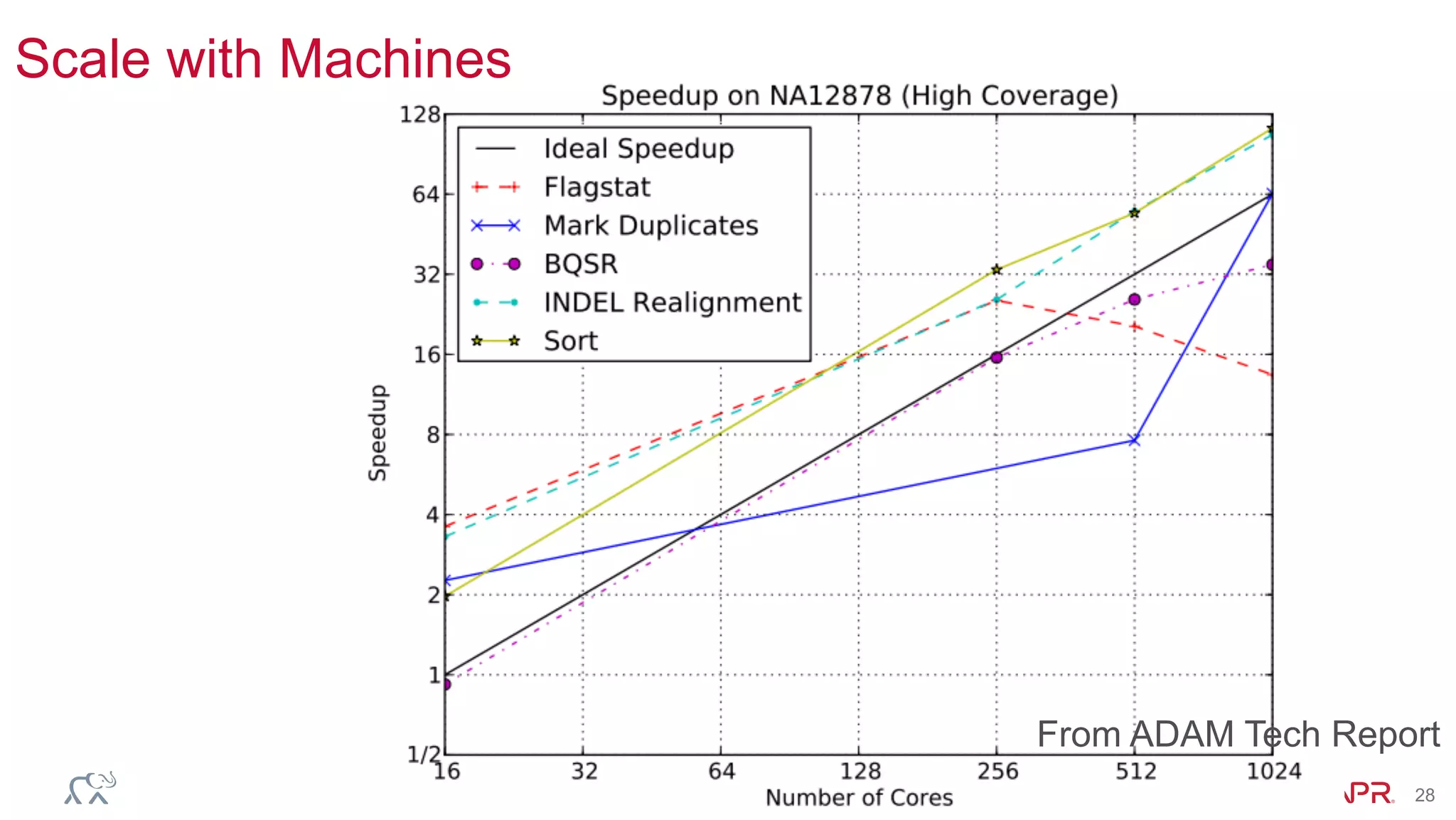
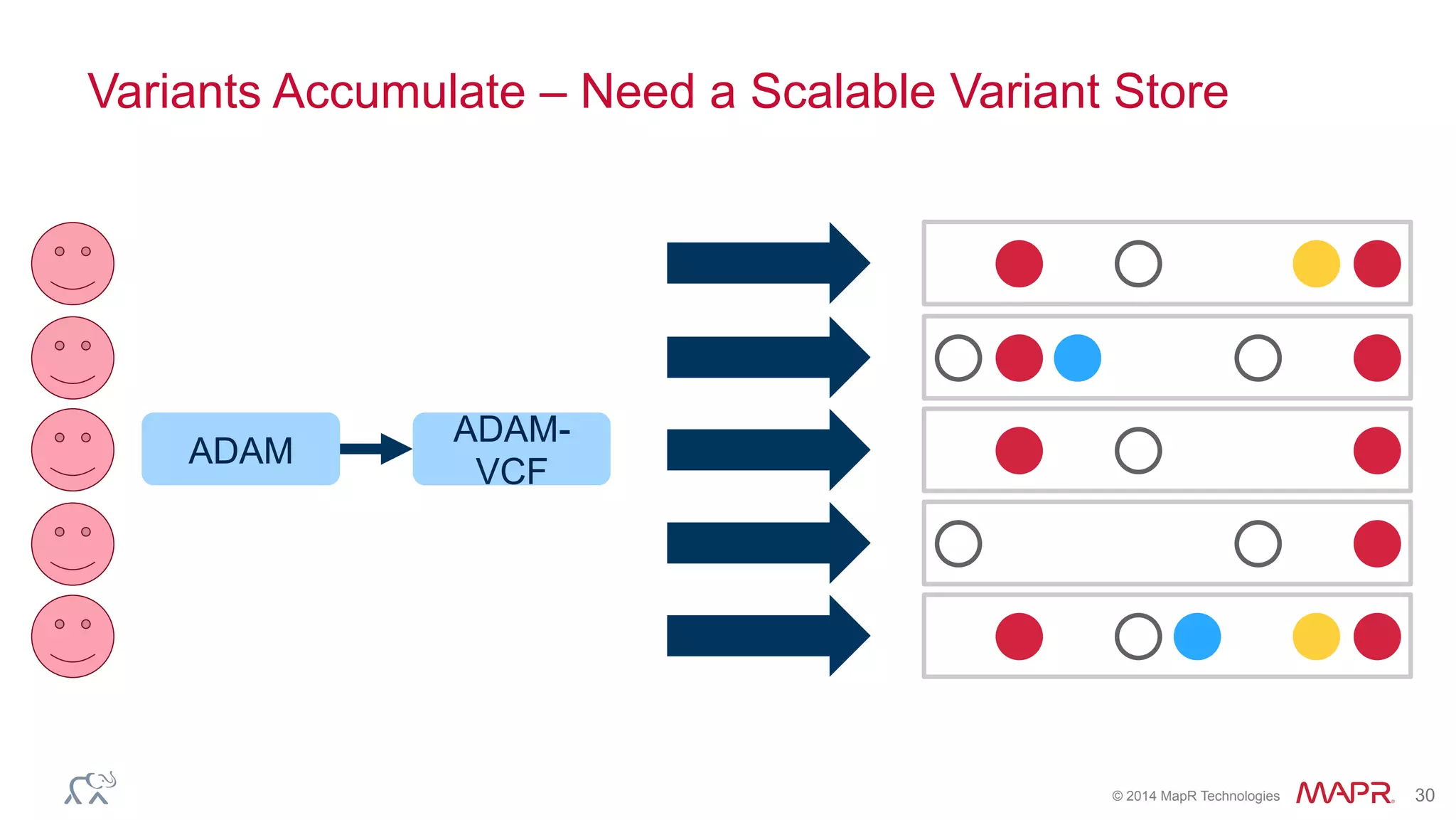
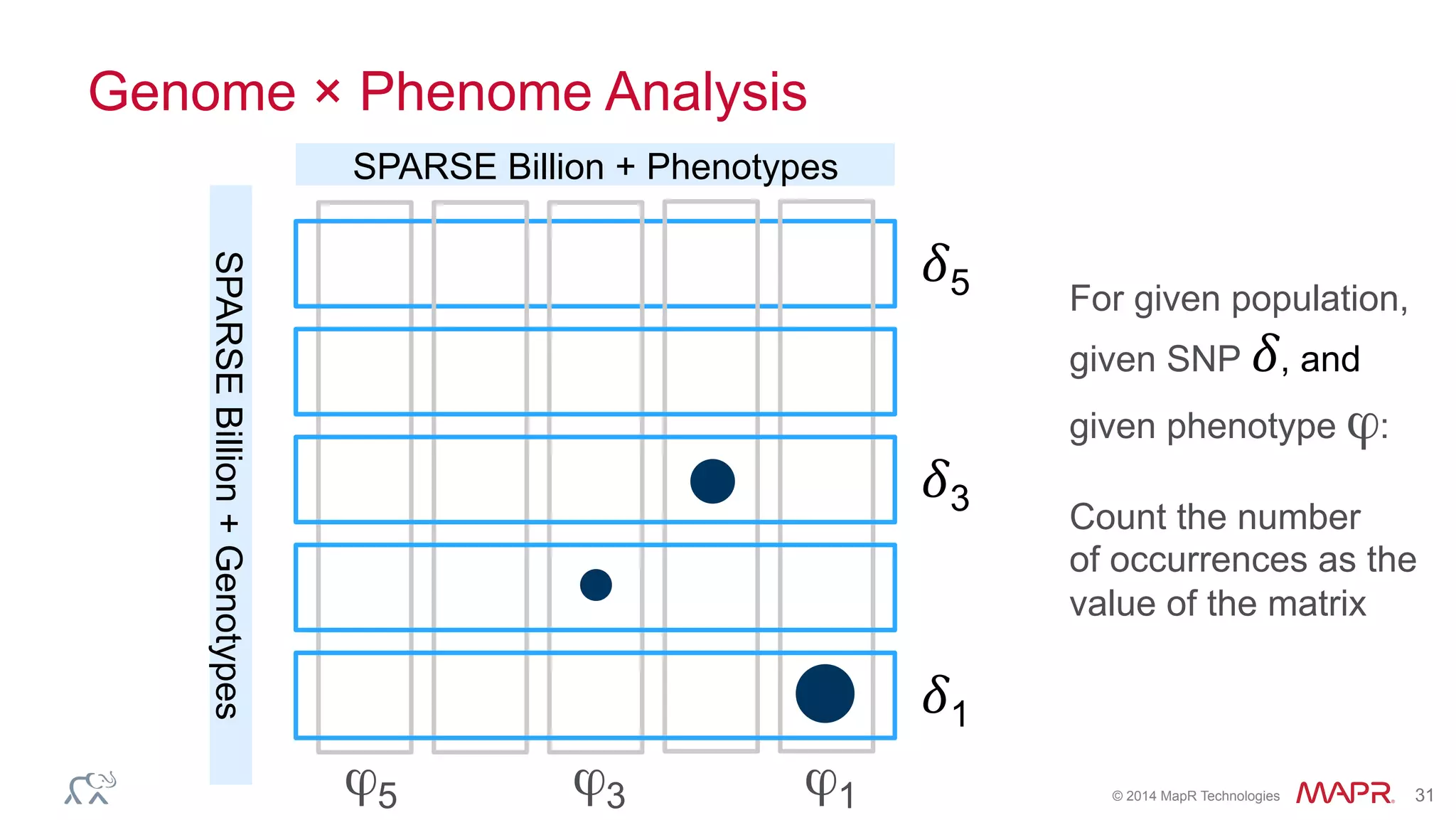
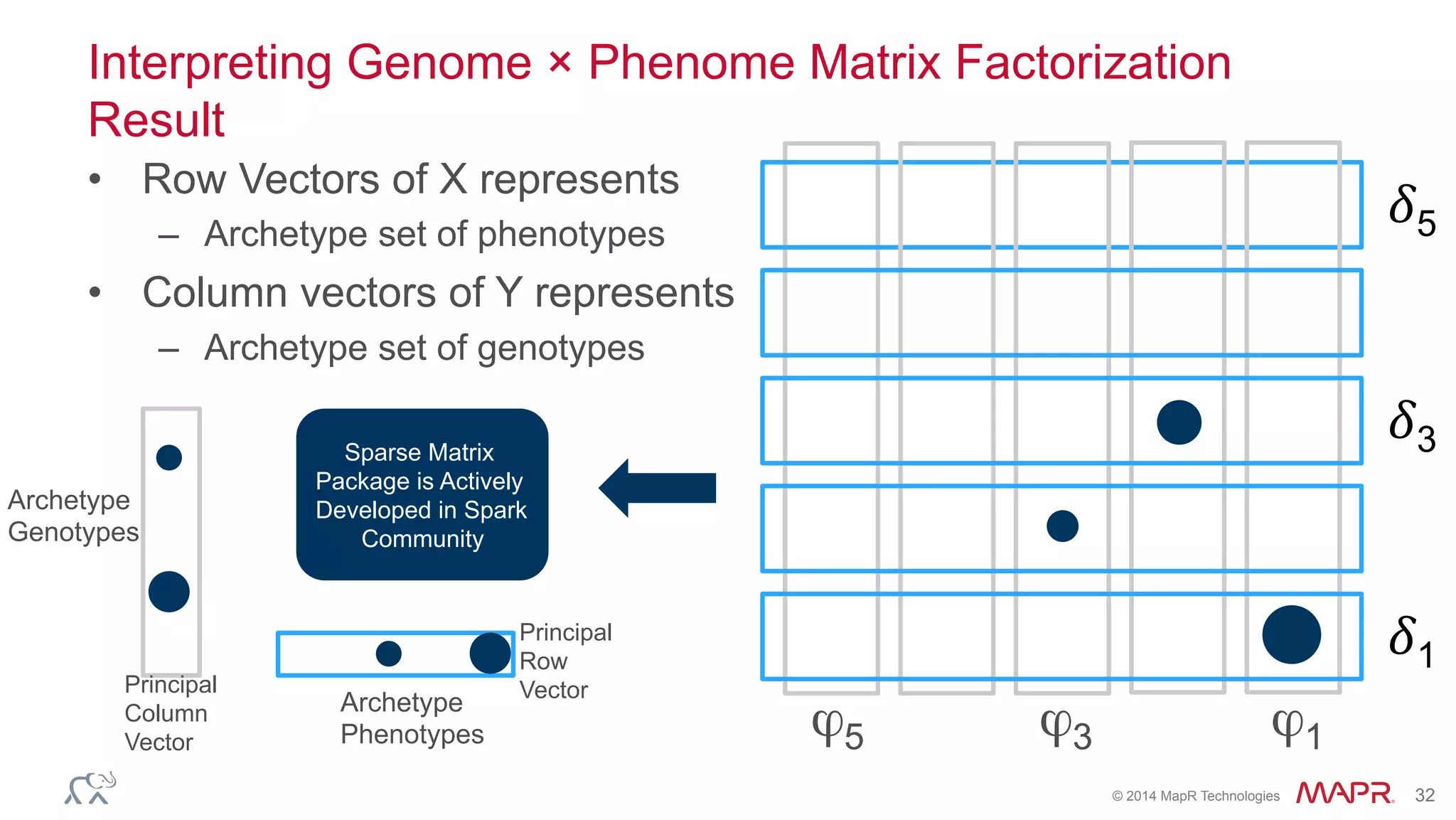
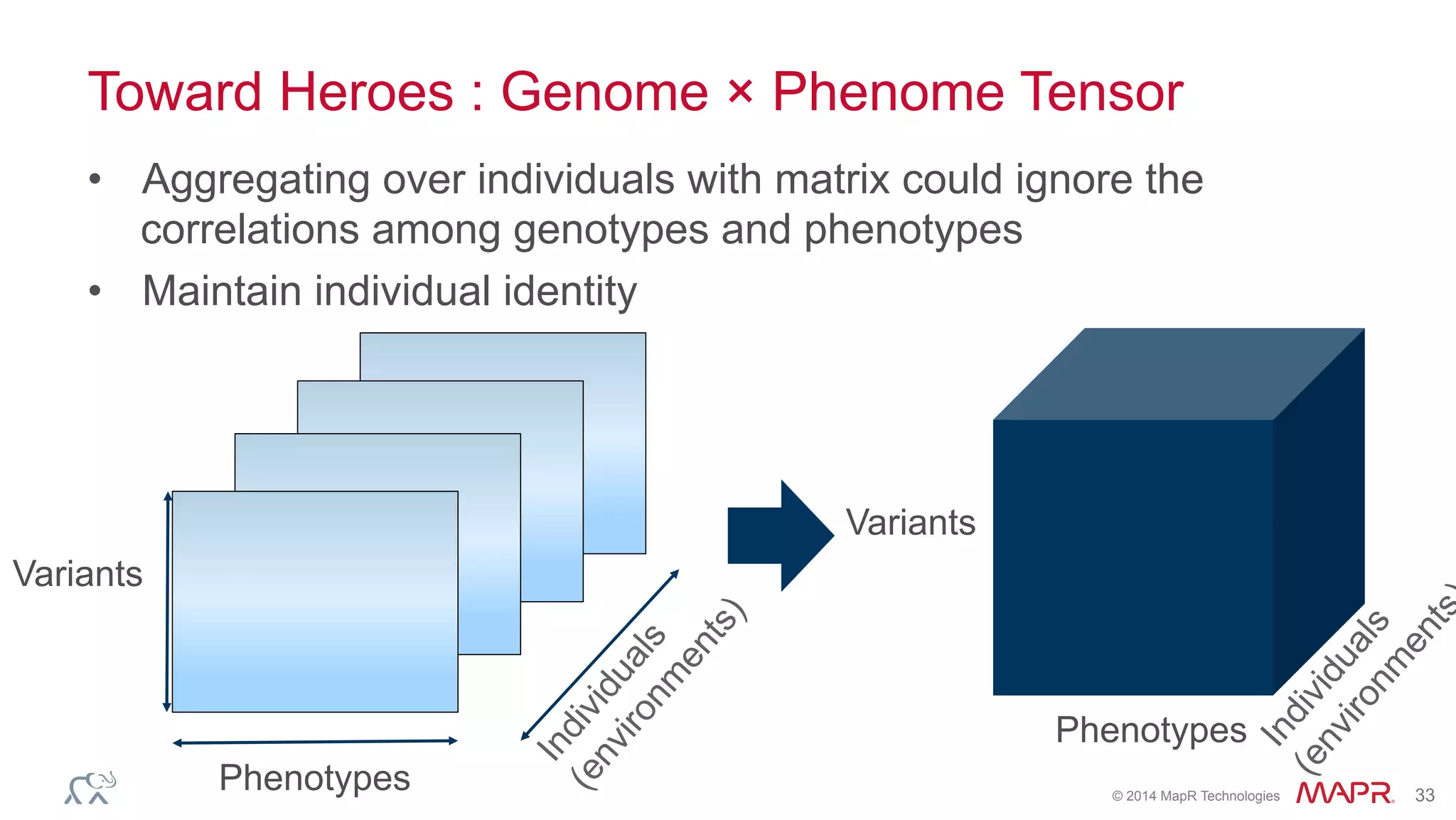
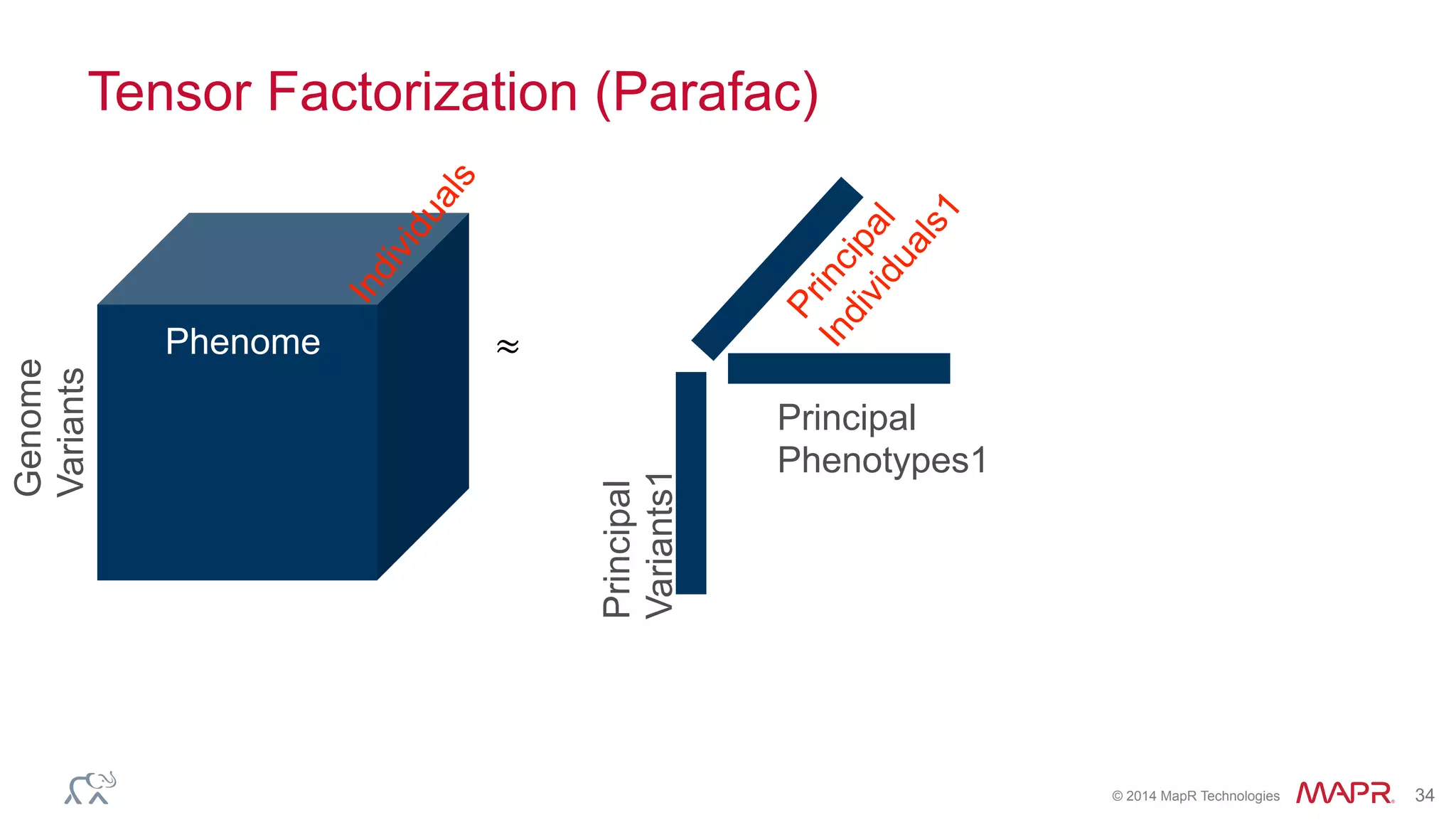
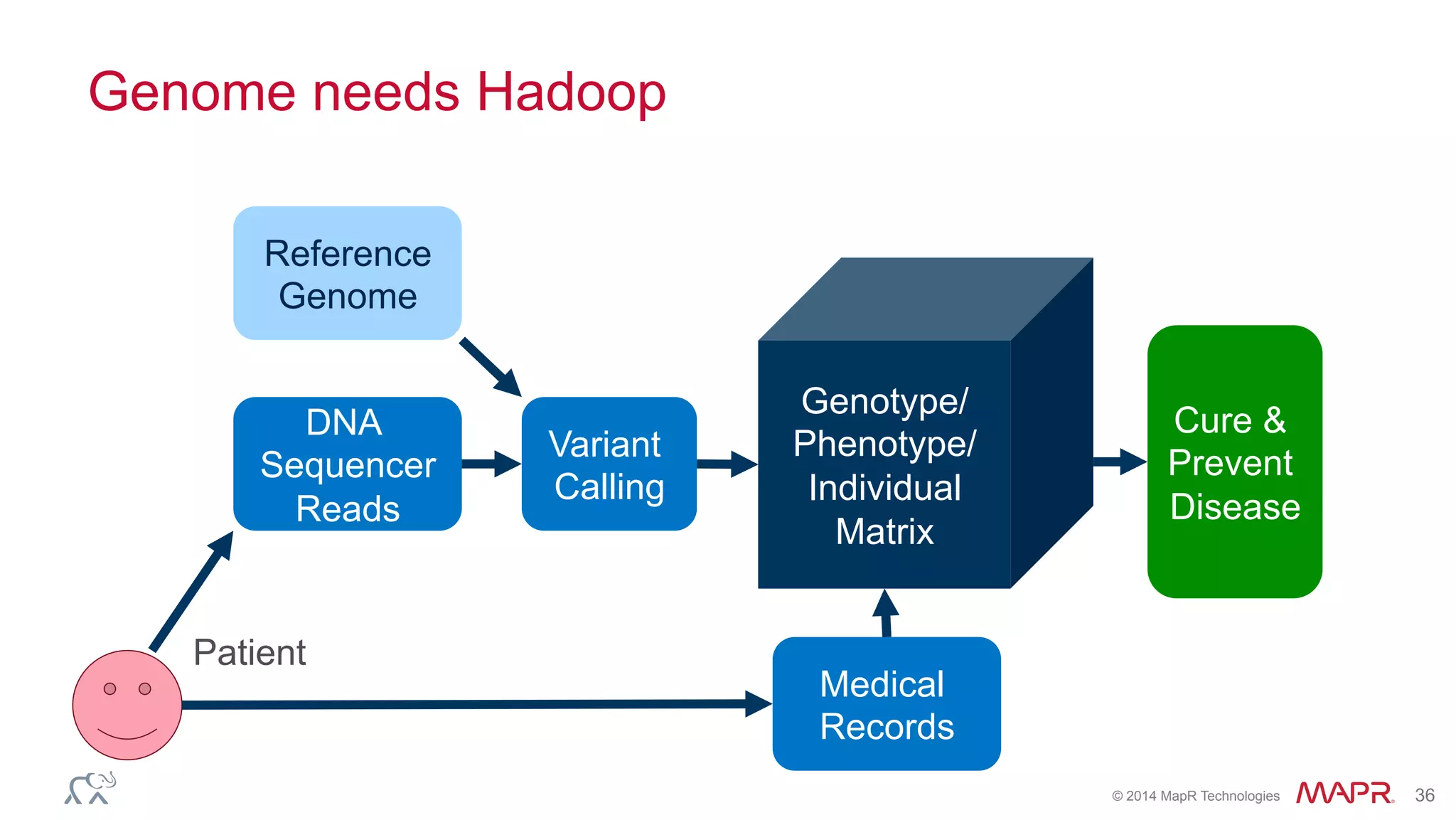
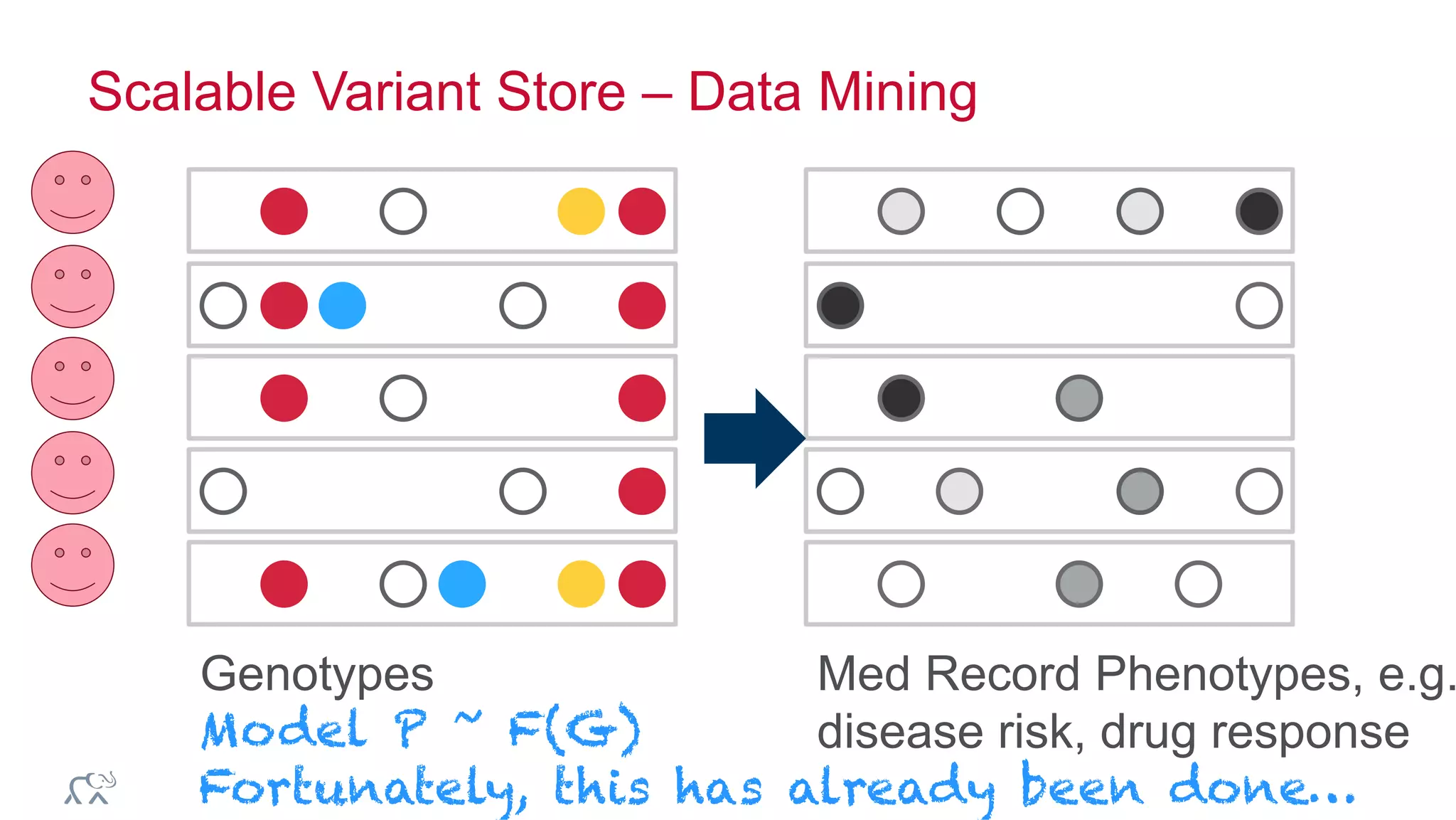
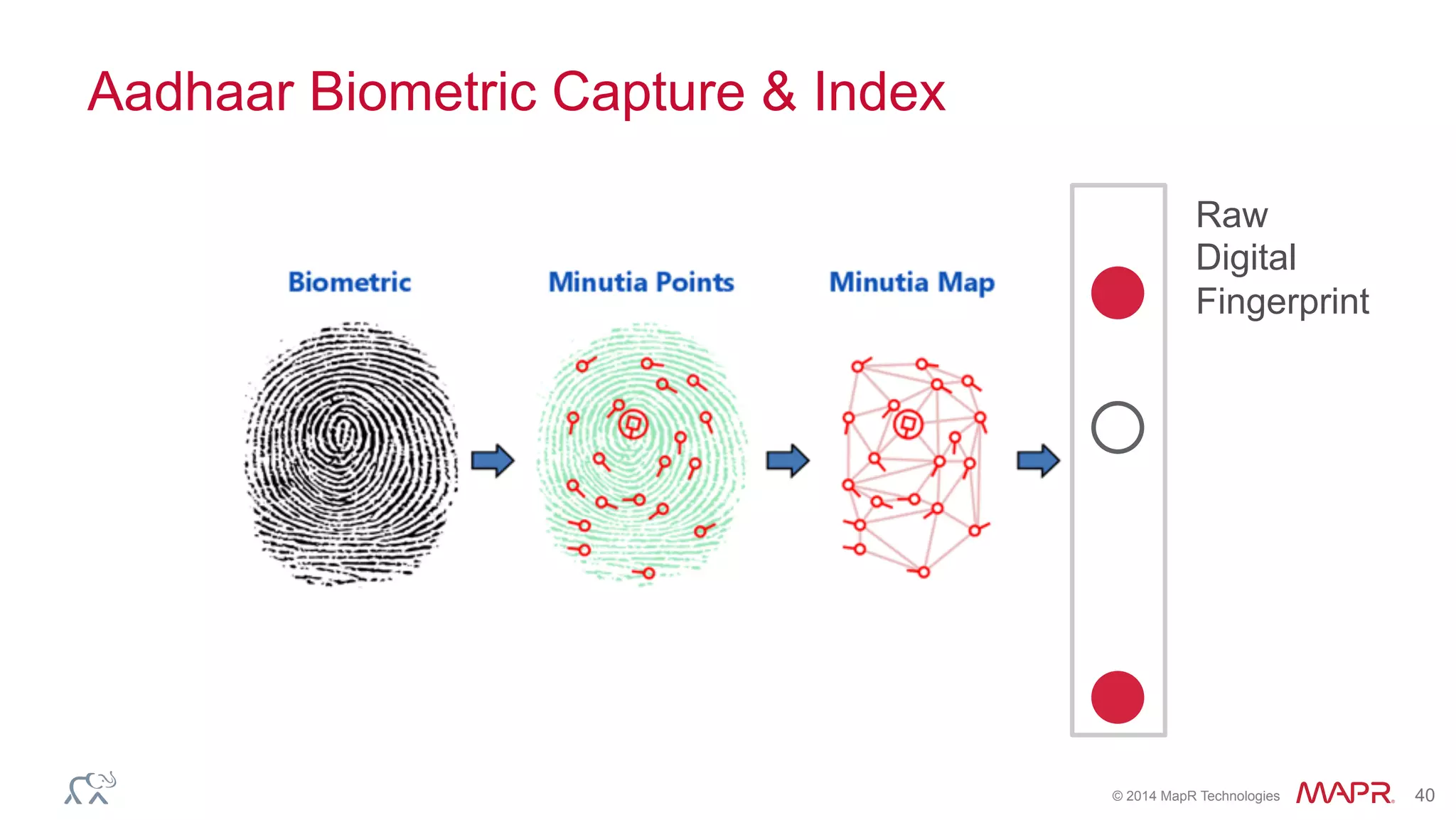
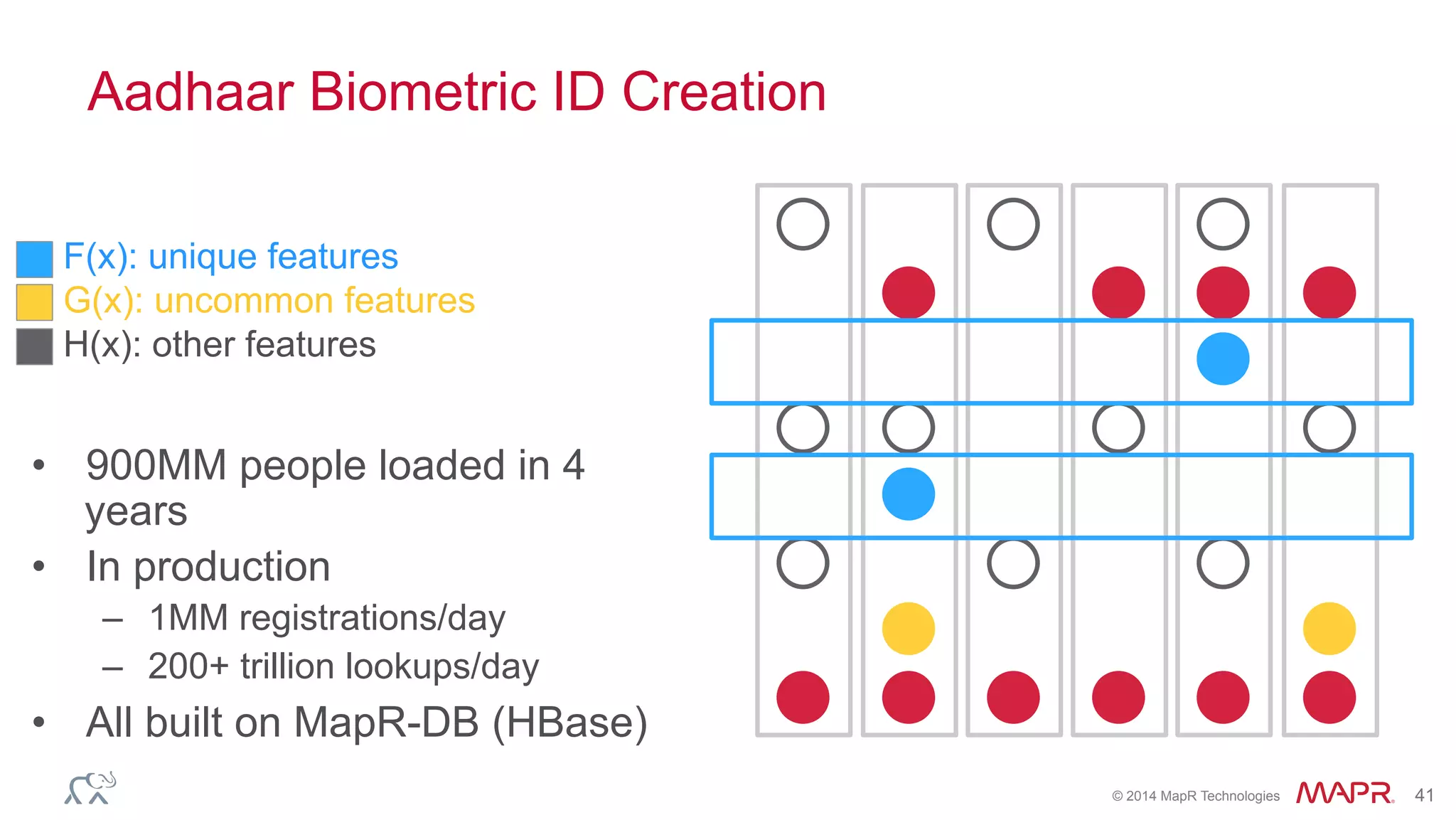
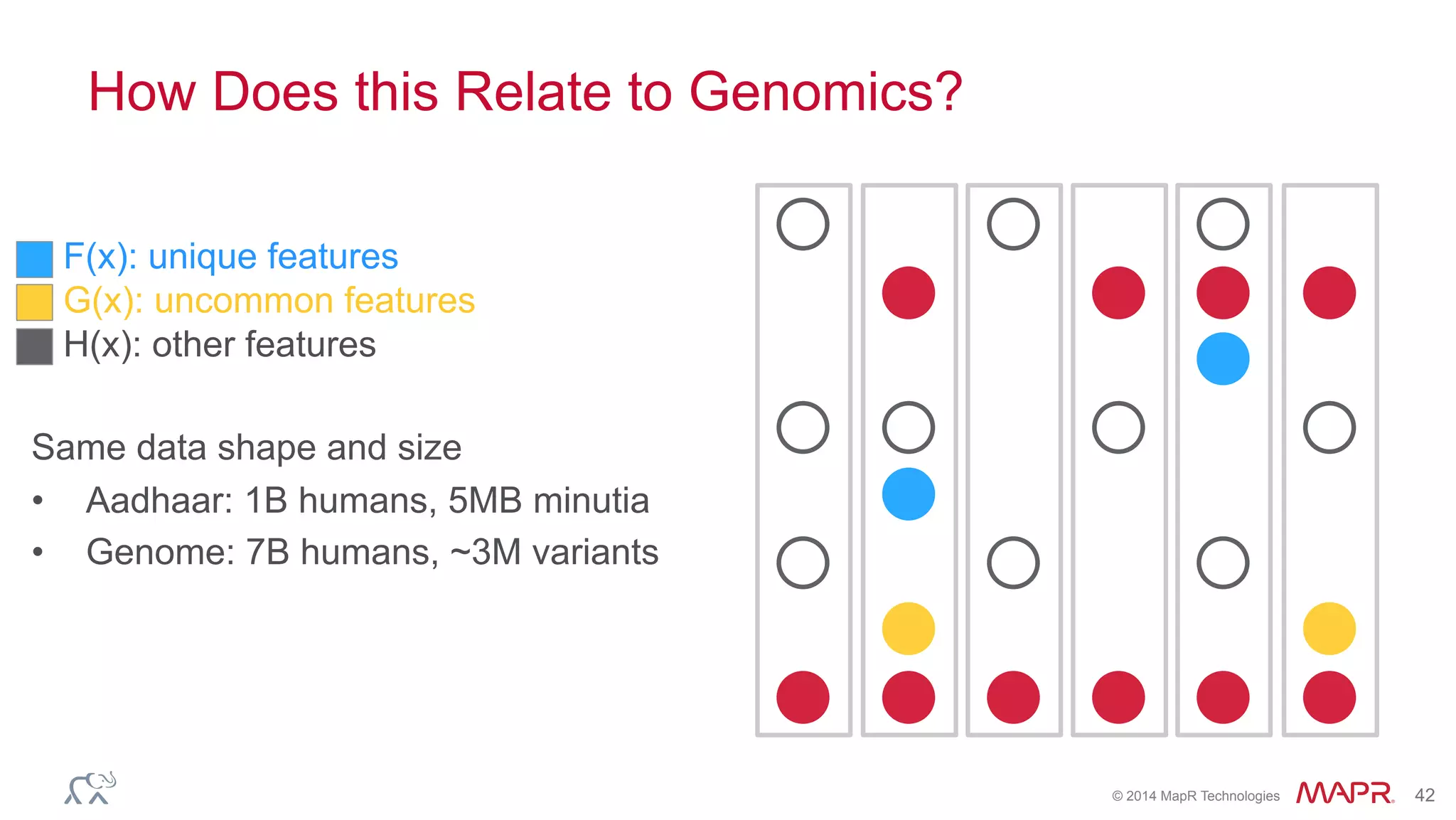
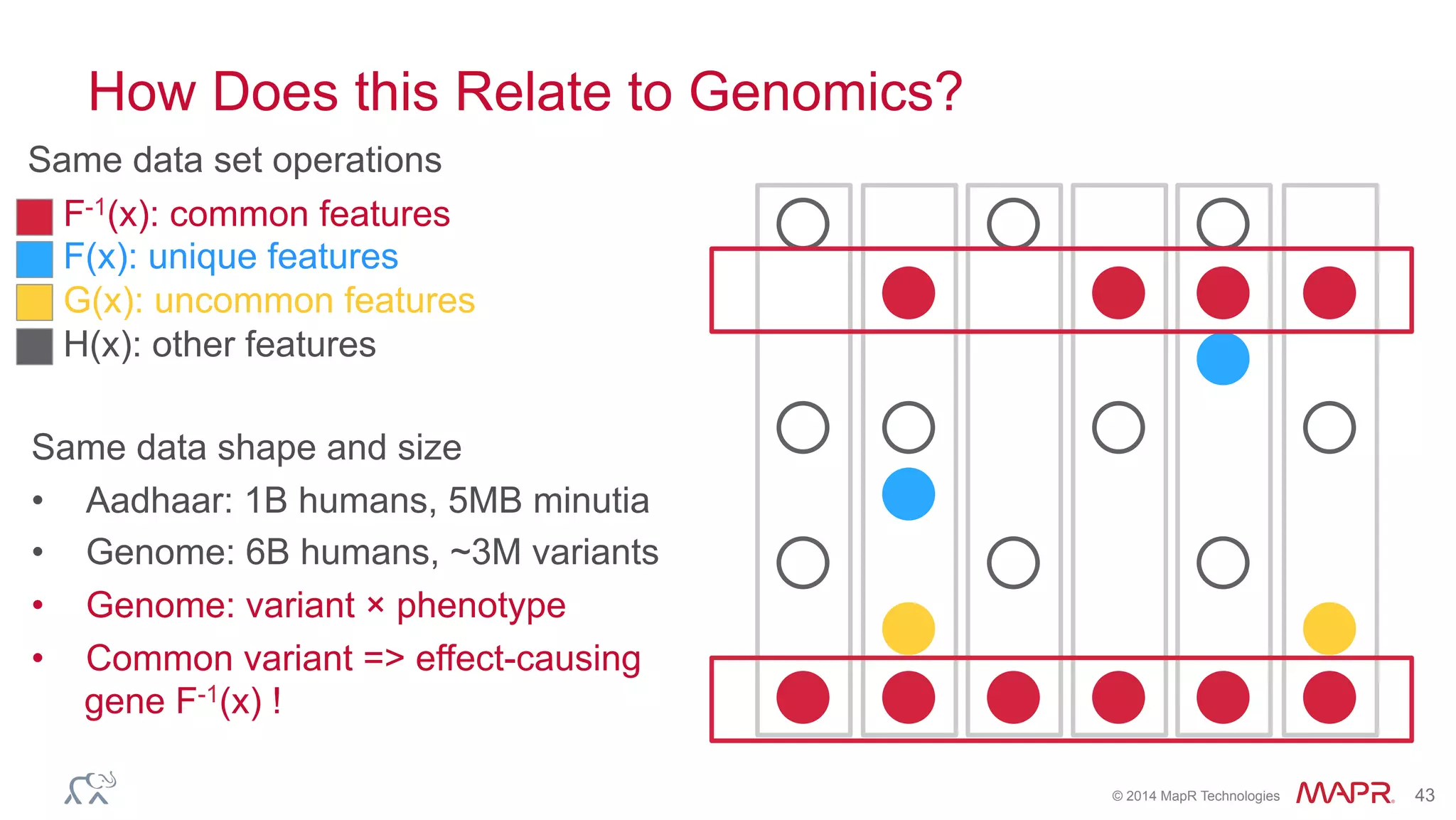
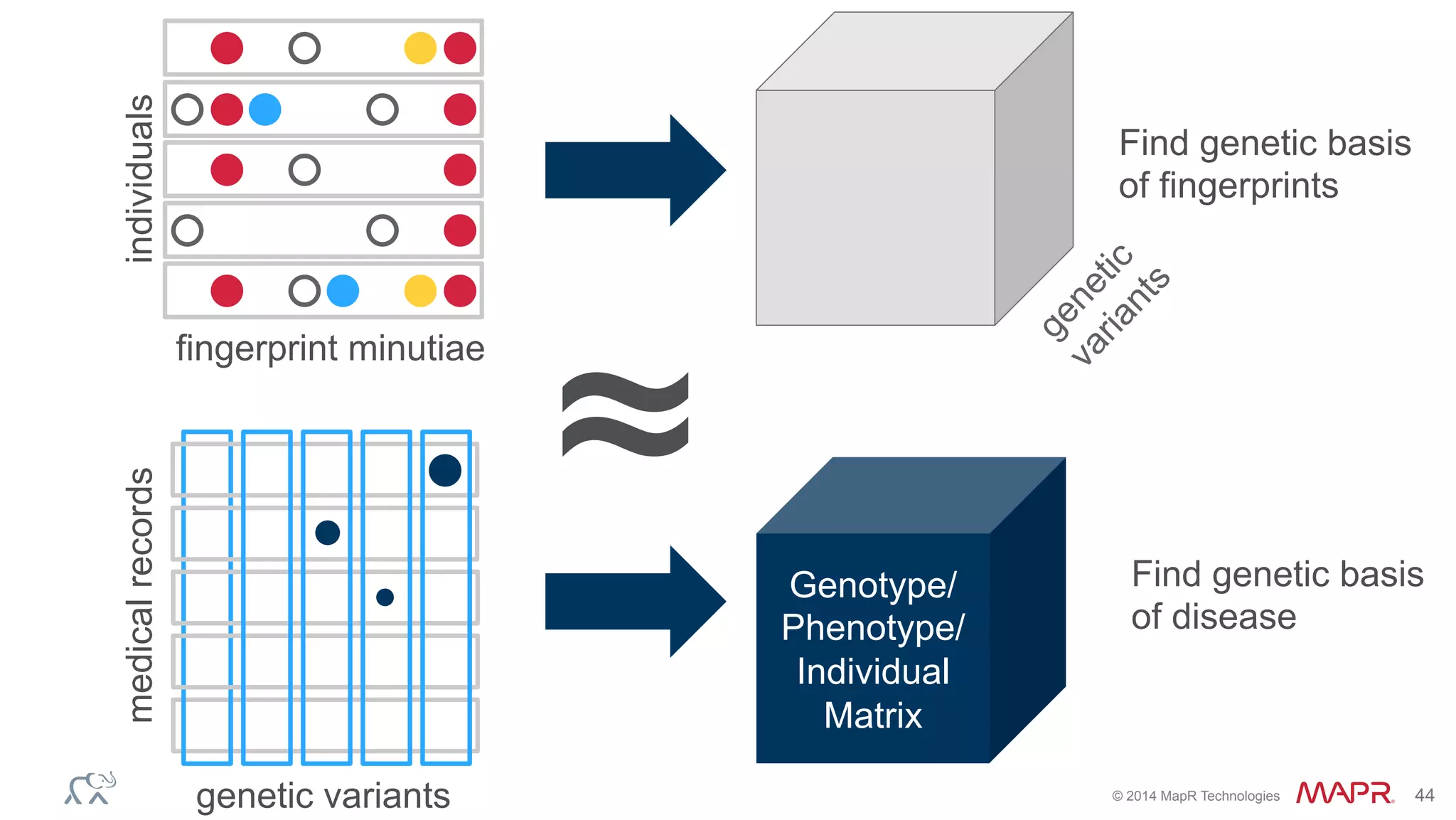
The document discusses the evolution of DNA sequencing and its implications for genomics, emphasizing the shift from research-focused applications to clinical analytics and personalized medicine. It highlights various analytics applications, including genomic data analysis and forensic applications, while addressing the need for scalable processing solutions beyond traditional Hadoop. Additionally, the document references India's Aadhaar initiative as an example of leveraging biometric data management in large-scale identity verification, drawing parallels to genomic data processing.