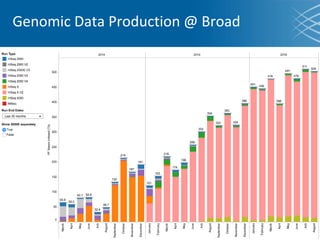
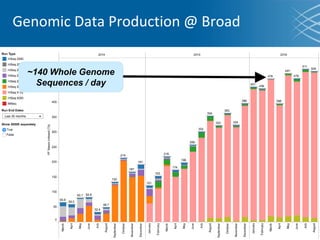
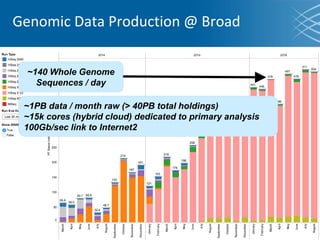
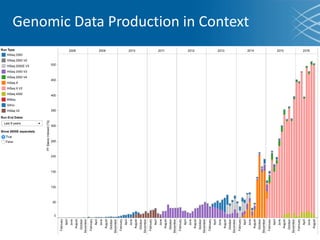
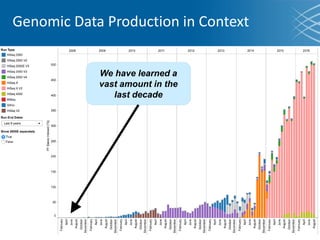
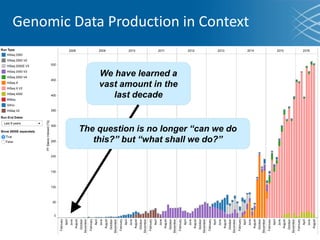
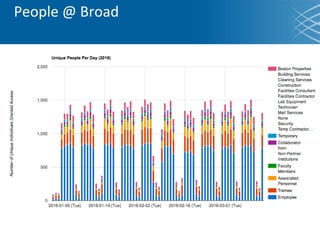
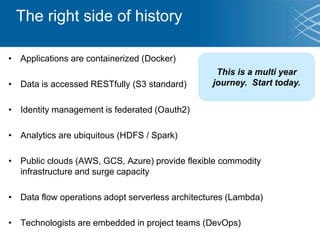
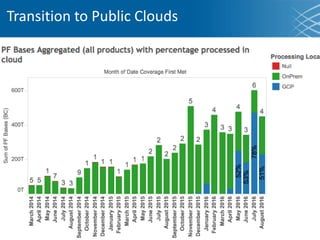
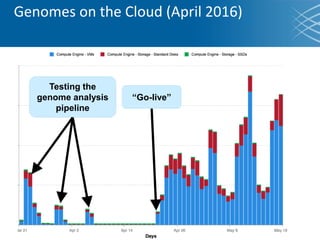
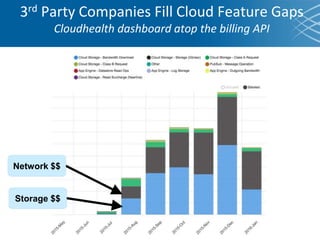
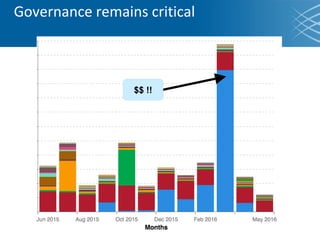
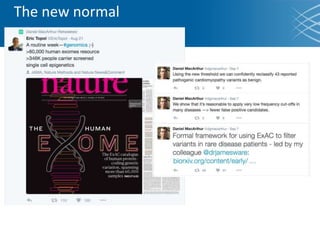
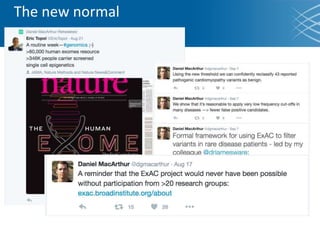
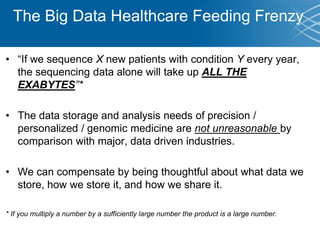
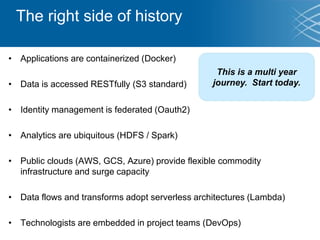
The document discusses the early development of genomic medicine and emphasizes the importance of collaboration, data analysis, and addressing privacy and security challenges. It highlights the Broad Institute's capabilities in genomic data production and the necessity for innovative technology and practices for effective data sharing and usage. The need for regulatory reforms and incentive structures to promote accessible and useful data is also underscored as organizations prepare for the future of genomic medicine.