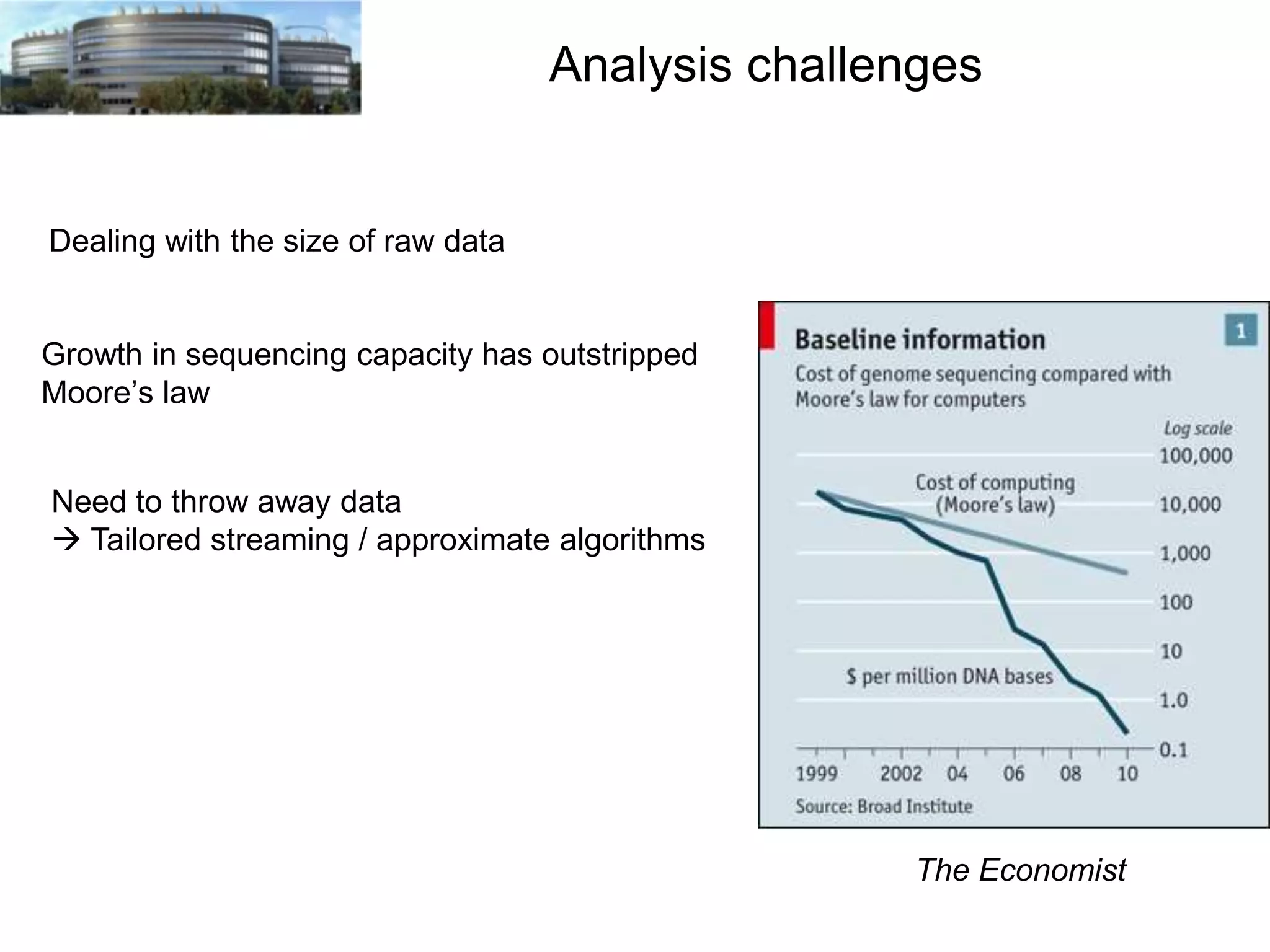
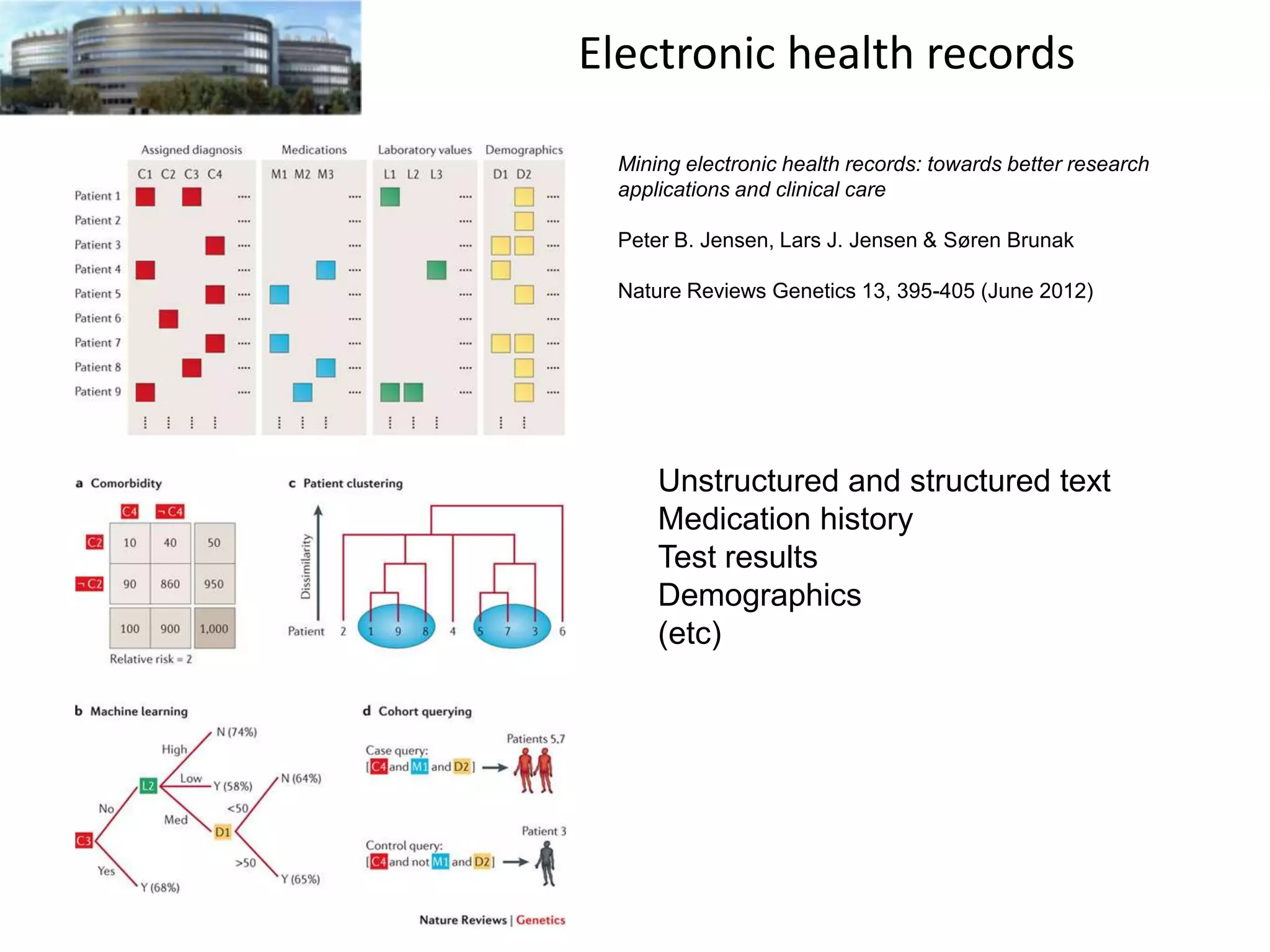
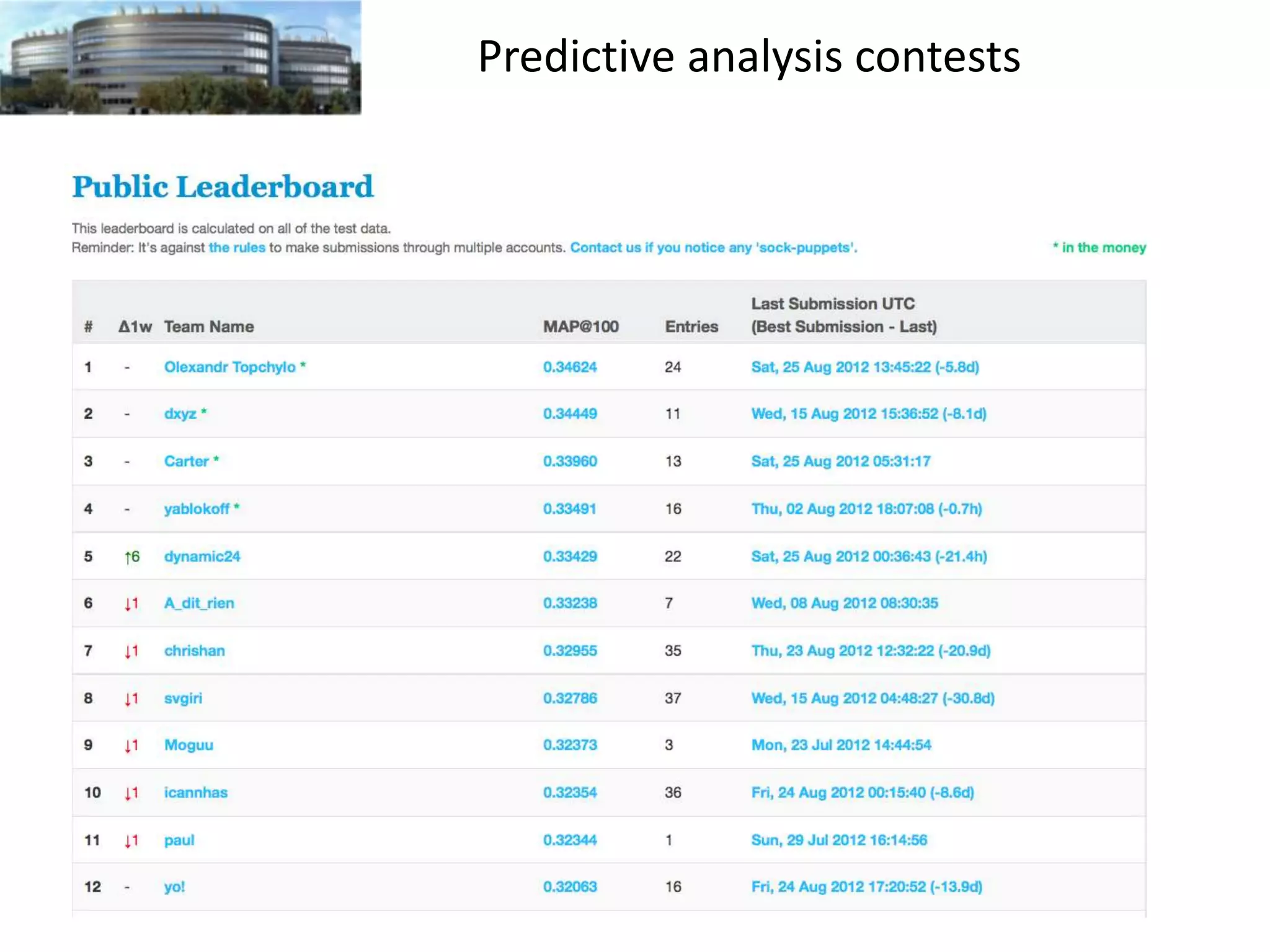
The document discusses the challenges and applications of data analysis in genomics, highlighting the rapid advancements in DNA and RNA sequencing technologies at Science for Life Laboratory, Stockholm. It covers the goals of precision medicine, understanding natural diversity, and complex diseases, alongside the technical challenges of data storage, transfer, and analysis in big genomics data. The document also emphasizes the need for improved infrastructure and tools to effectively manage and interpret the increasing volume of genomic data.

















![Storage and transfer
“European Bioinformatics Institute (EBI) stores 20 pb of data, of which 2 pb is
genomic”
“Single human genome ~140 Gb”
“ … downloading the data is time-consuming, and researchers must be sure that their
computational infrastructure and software tools are up to the task. “If I could, I would
routinely look at all sequenced cancer genomes,” says [Arend] Sidow. “With the
current infrastructure, that's impossible.”
Cloud solutions:
Embassy Cloud – EBI + CSC in Espoo
easyGenomics – BGI Hong Kong
DNANexus – commercial service, Silicon Valley](https://image.slidesharecdn.com/datamininguppsala-131213164622-phpapp01/75/Data-analytics-challenges-in-genomics-32-2048.jpg)