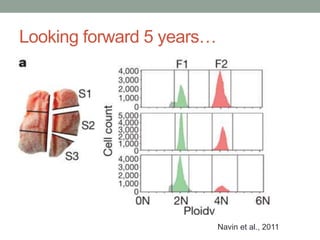
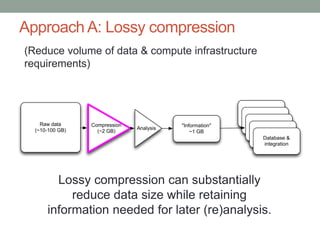
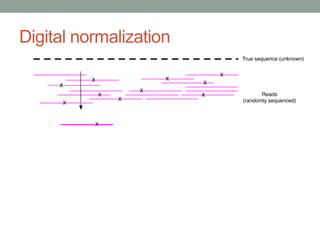
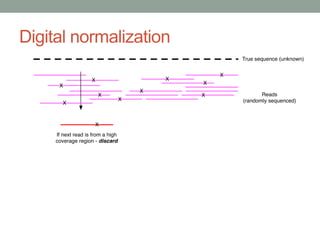
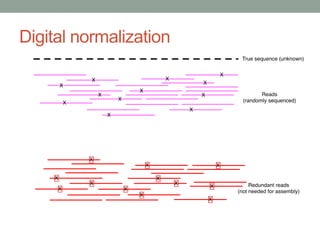
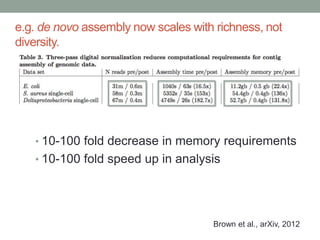
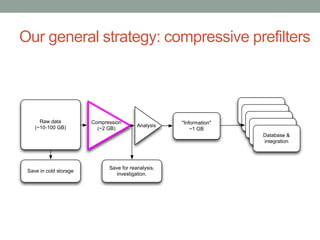
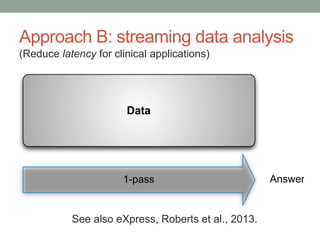
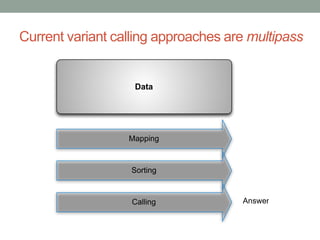
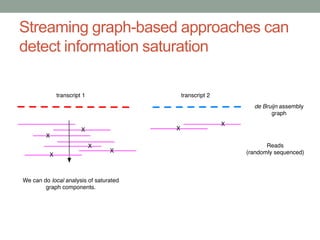
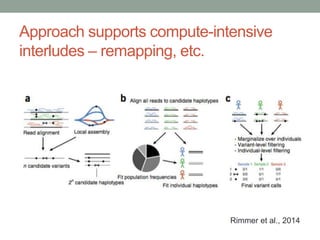
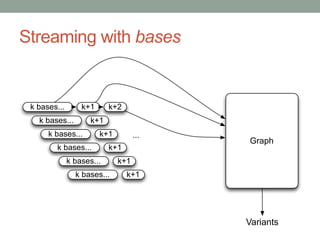
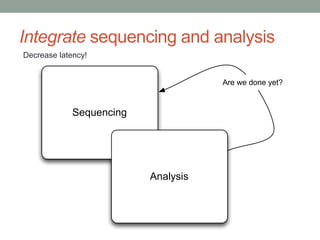
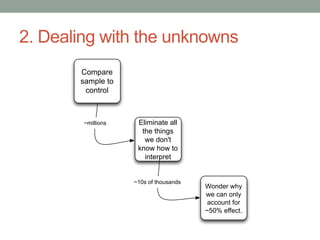
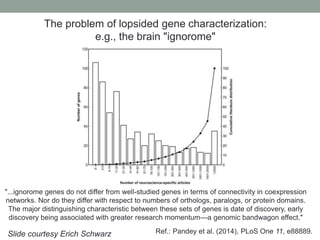
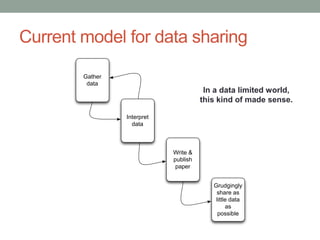
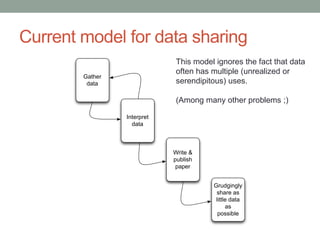
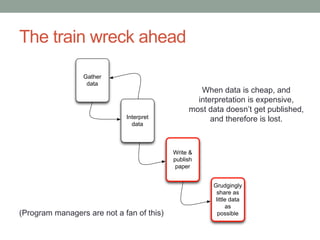
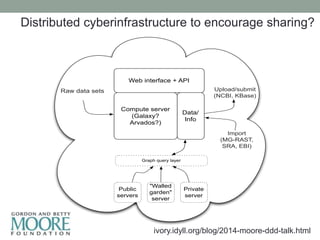
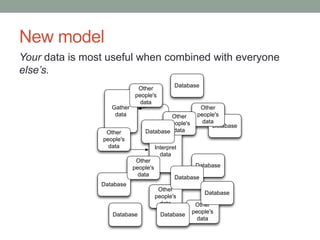
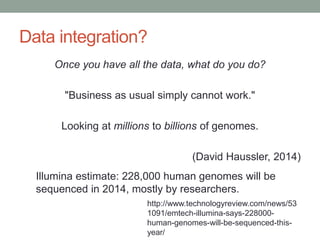
This document discusses the challenges and opportunities presented by the increasing volume and complexity of biological data. It outlines four main areas: 1) Developing methods to efficiently store, access, and analyze large datasets; 2) Broadening our understanding of gene function beyond a small number of well-studied genes; 3) Accelerating research through improved sharing of data, results, and methods; and 4) Leveraging exploratory analysis of integrated datasets to generate new insights. The author advocates for lossy data compression, streaming analysis, preprint sharing, improved metadata collection, and incentivizing open data practices.