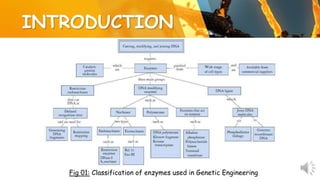
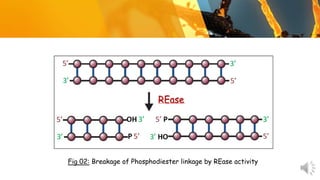
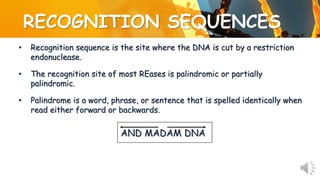
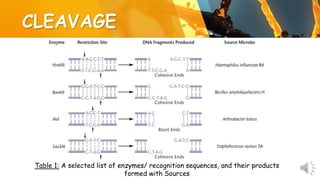
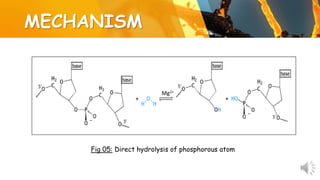
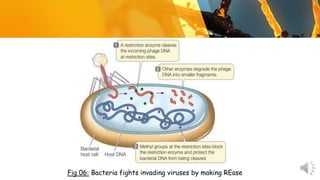
This document presents information about restriction endonucleases. It begins with an introduction and history, then discusses the nomenclature, recognition sequences, and classification of restriction endonucleases. It describes the four main types of restriction endonucleases, their subunit composition, cofactors used, characteristics, and mechanisms of action. The document concludes with references cited.