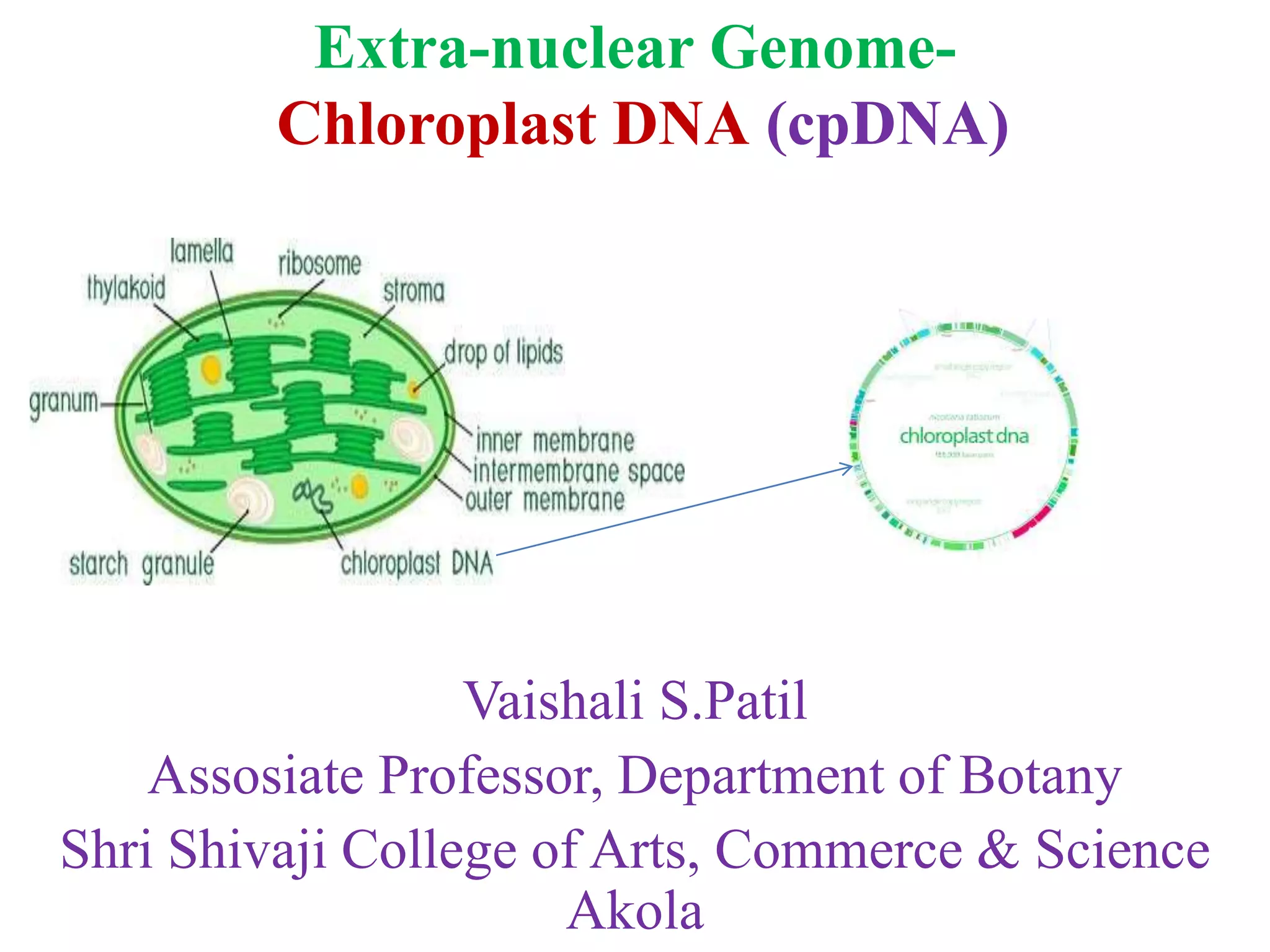
1. Chloroplast DNA (cpDNA) is circular DNA located in chloroplasts that contains genes essential for photosynthesis. These genes are inherited extra-nuclearly and do not follow Mendelian patterns of inheritance.
2. In 1909, Correns discovered that four o'clock plant leaf color was inherited maternally through the chloroplast rather than through nuclear genes. This was an early example of non-Mendelian cytoplasmic inheritance.
3. Chloroplast genes code for proteins involved in photosynthesis, though nuclear genes are also required. Mutations in chloroplast genes often result in white or yellow leaves due to disrupted chlorophyll production.