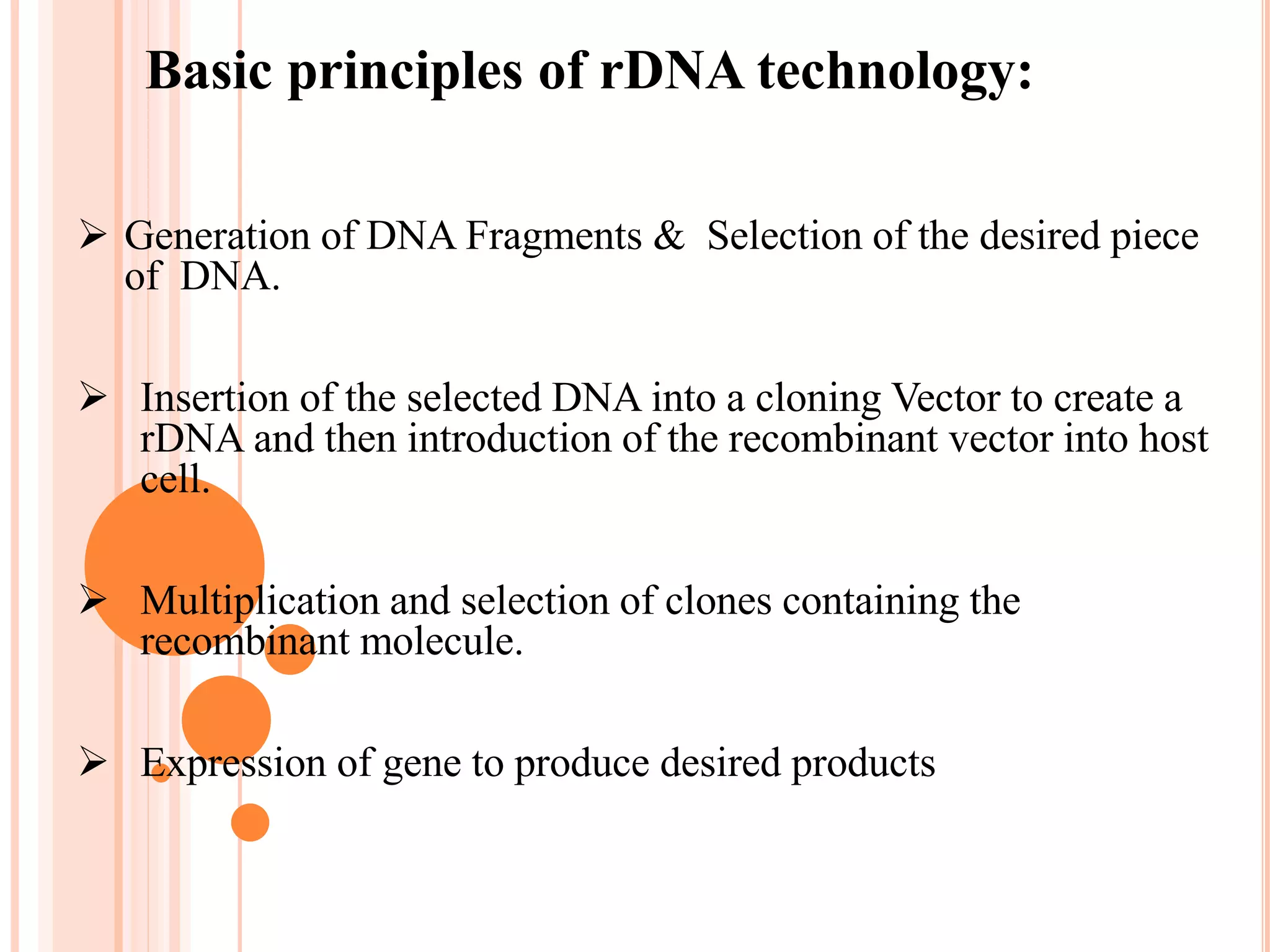
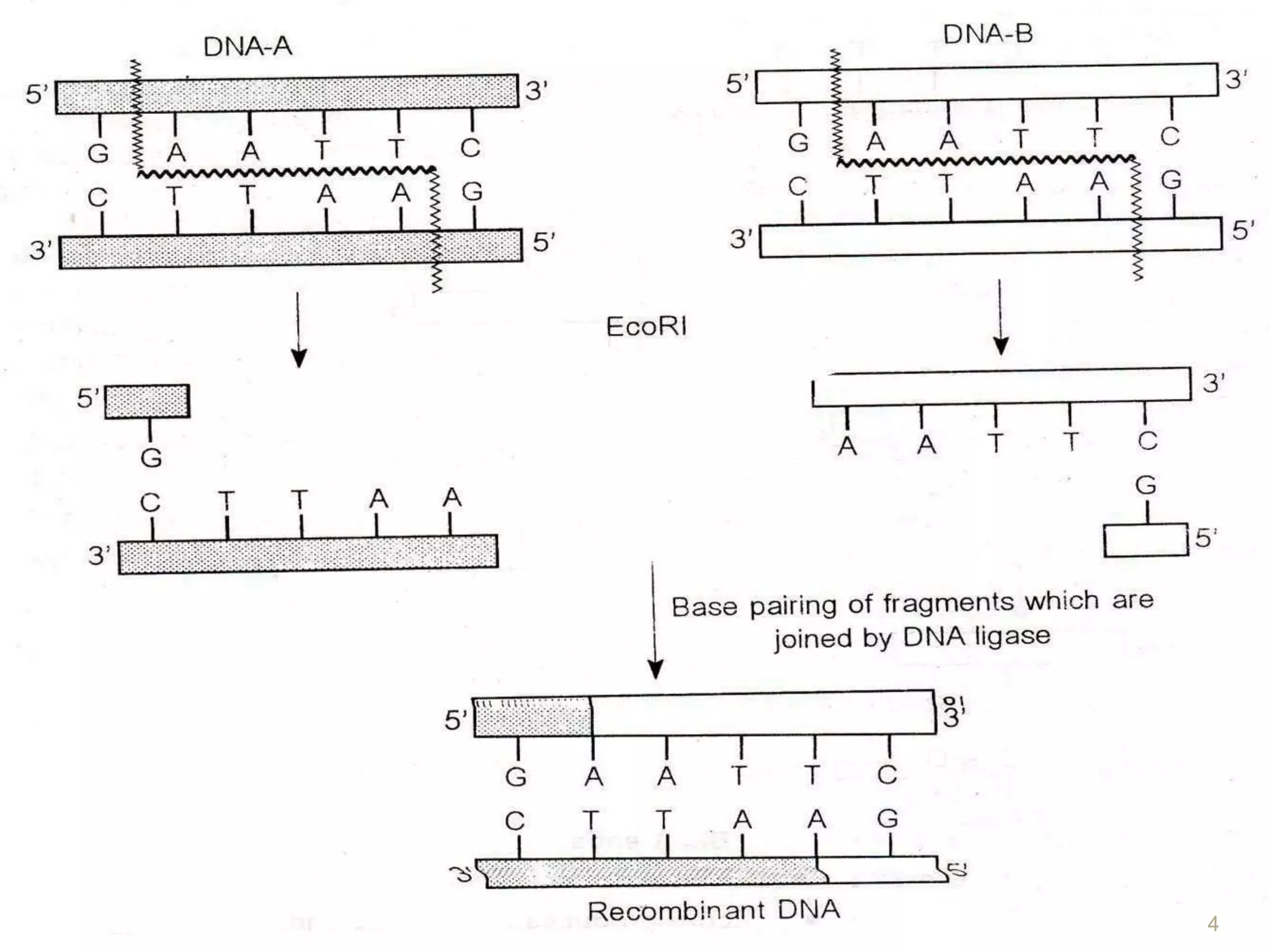
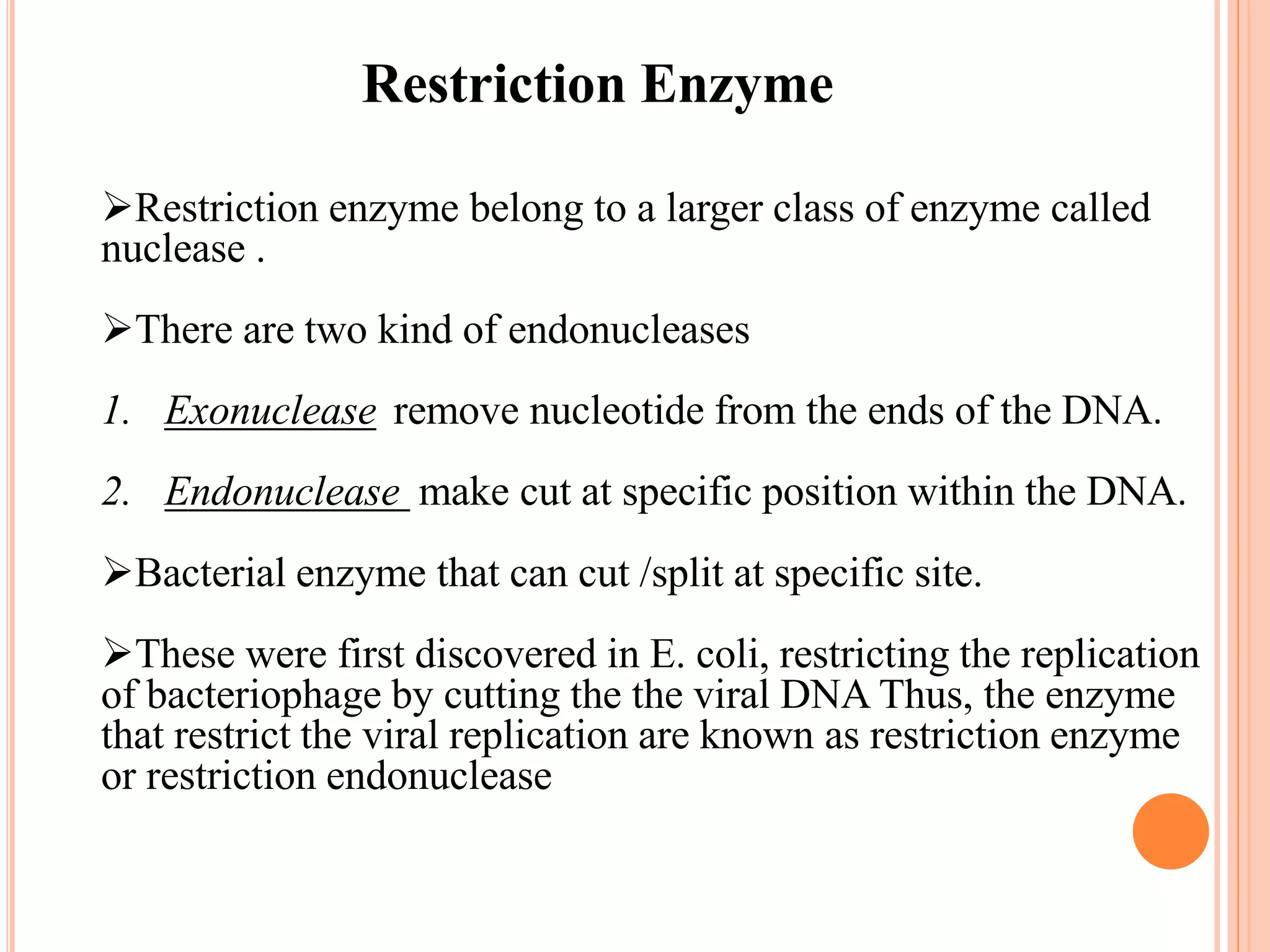
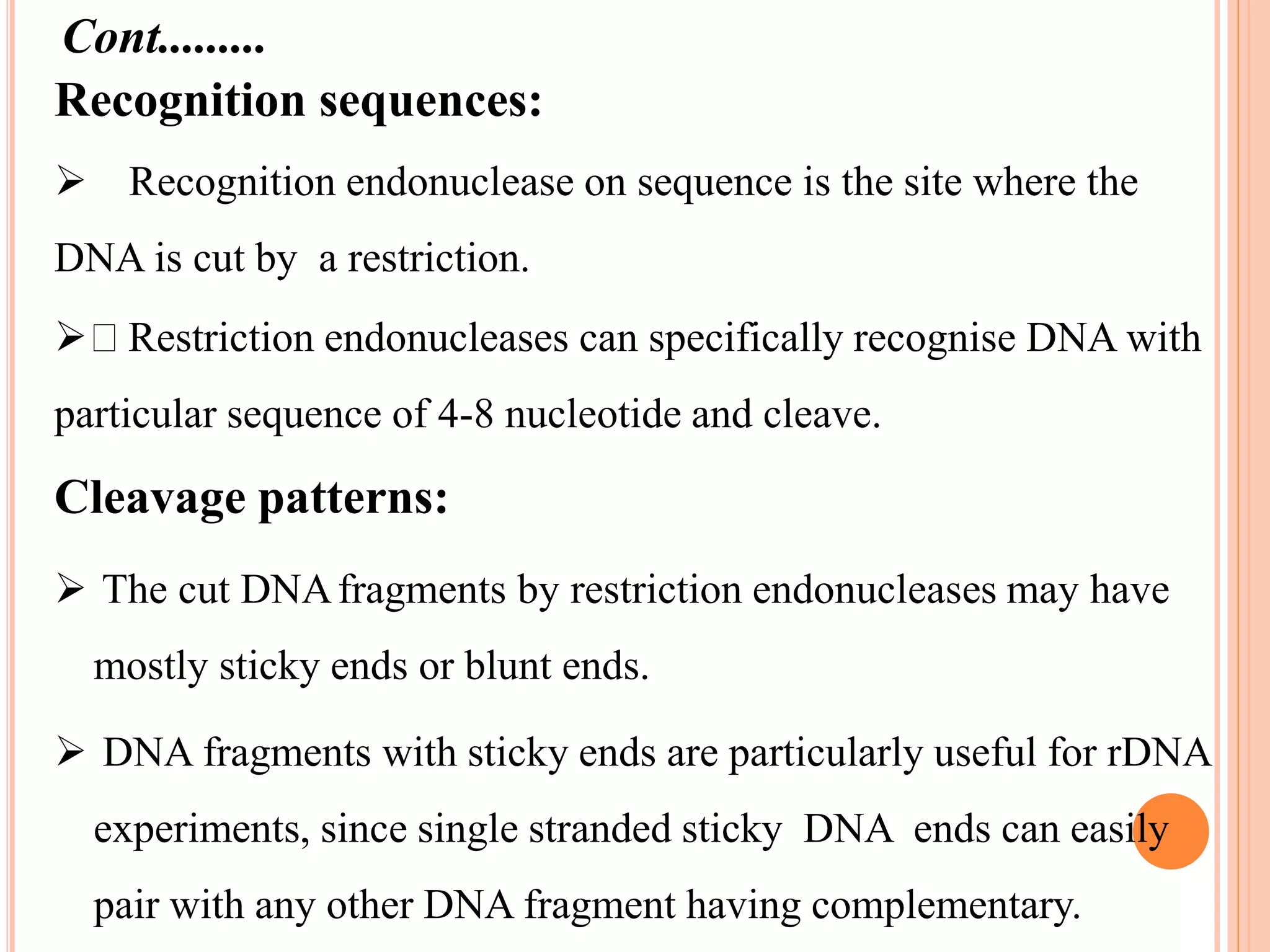
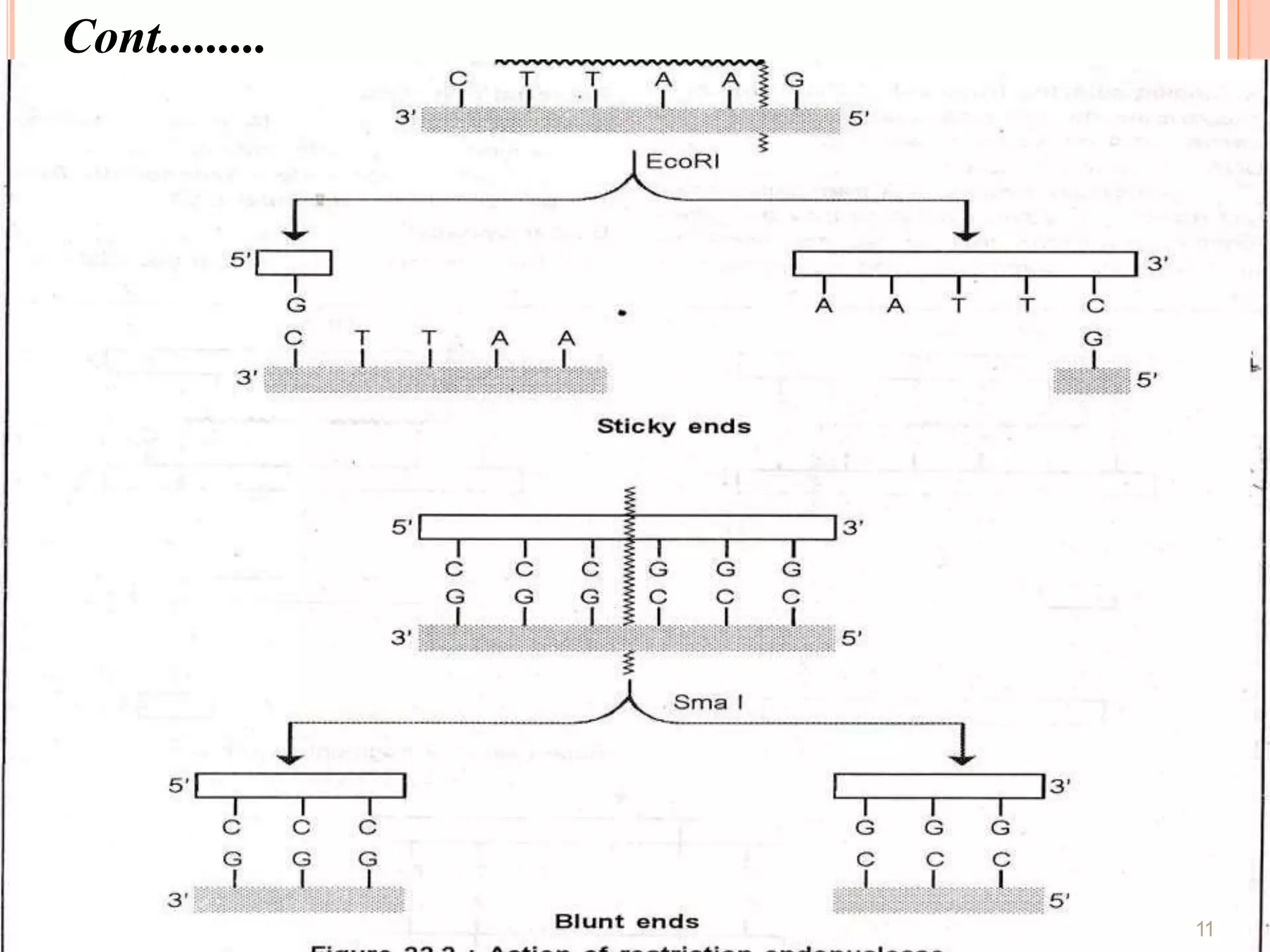
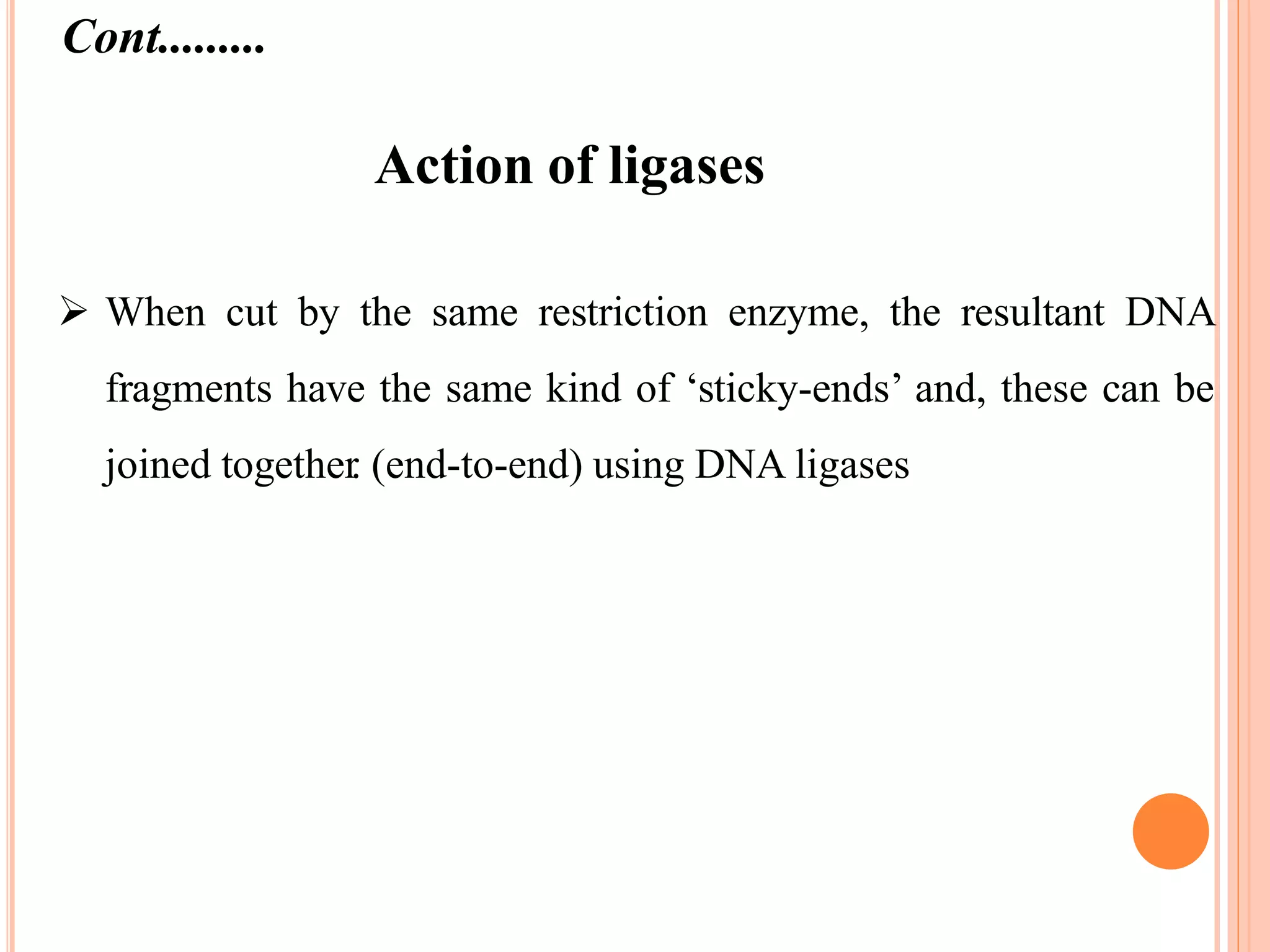
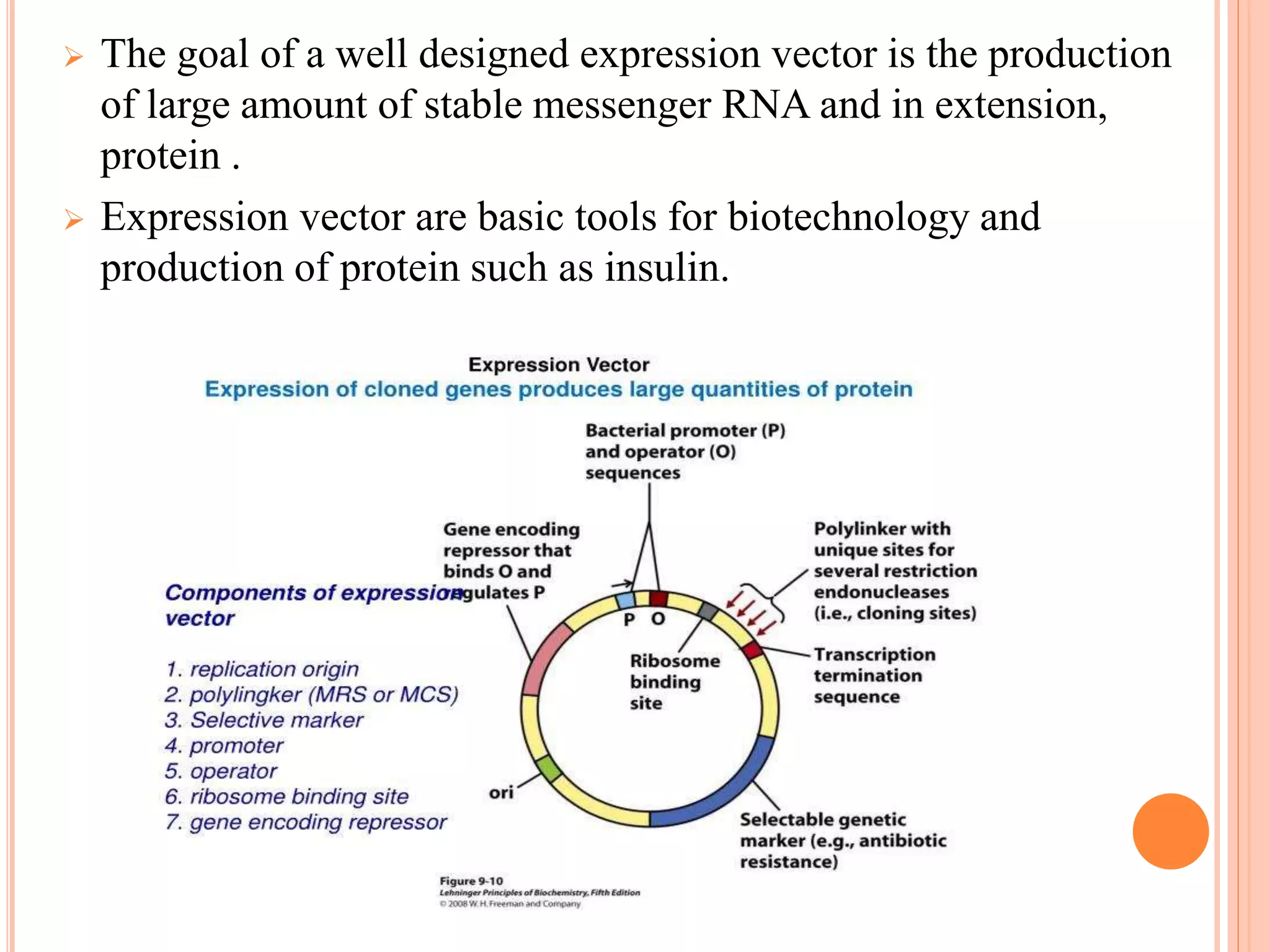
Recombinant DNA technology involves isolating, cutting, and splicing DNA from different species to create new recombinant molecules for practical applications. The core processes include generating DNA fragments, inserting them into cloning vectors, and multiplying the recombinant vectors within host cells to produce desired products. Key tools in this technology are restriction enzymes, ligases, and various vectors like plasmids and artificial chromosomes, all utilized in diverse fields such as agriculture, medicine, and forensic science.