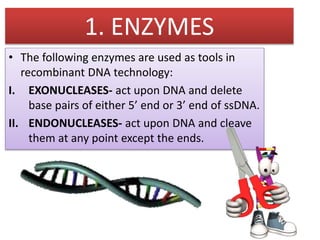
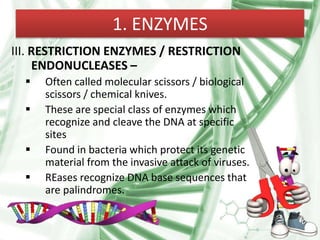
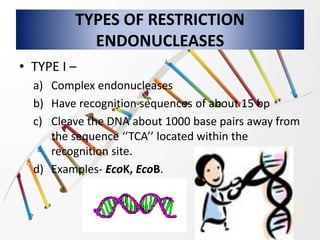
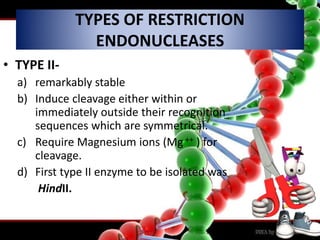
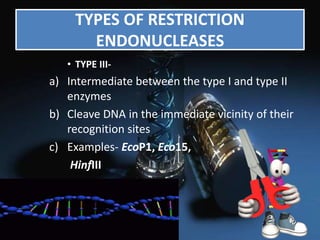
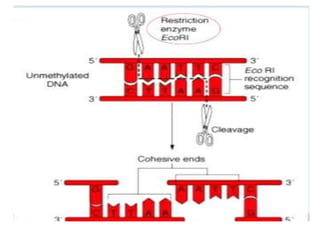
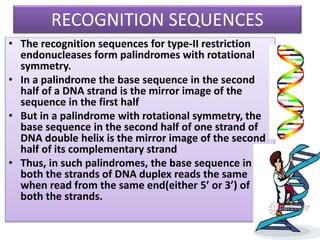
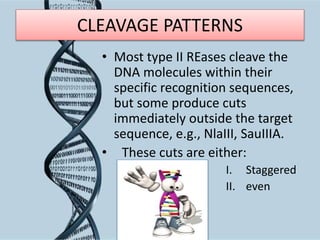
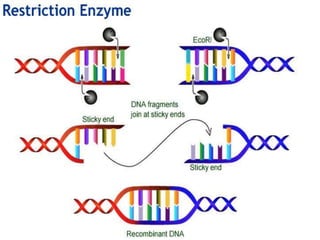
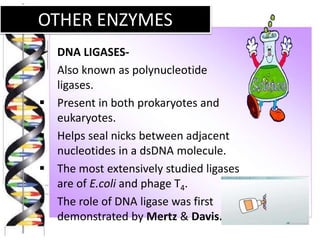
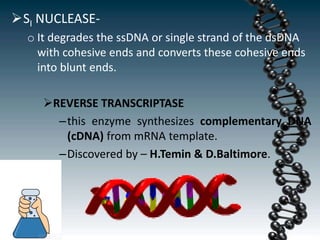
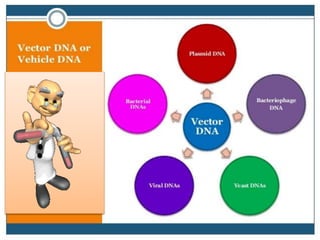
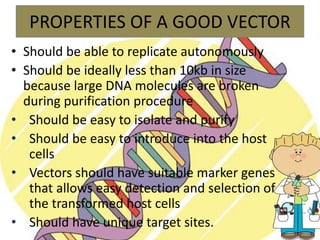
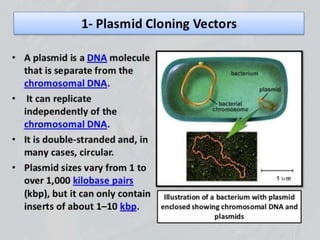
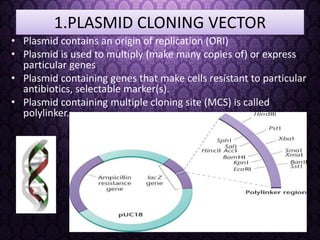
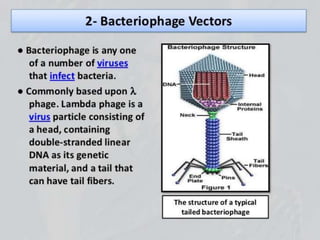
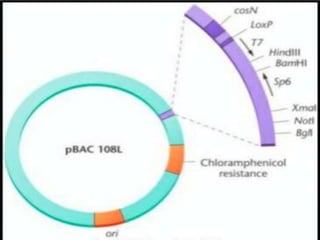
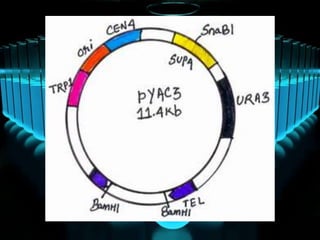
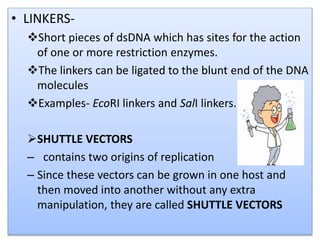
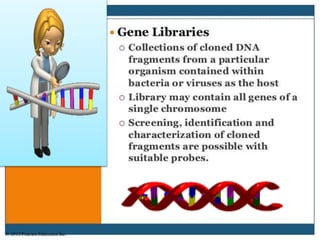
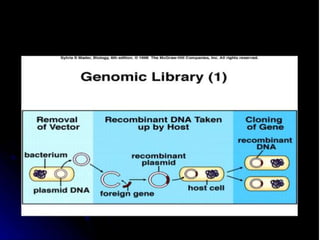
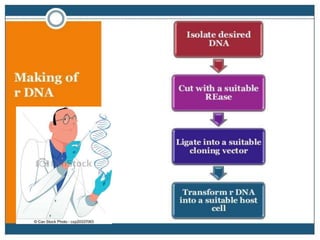
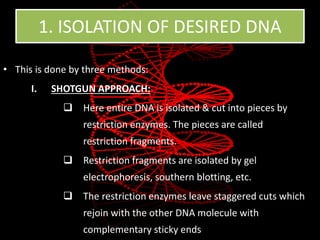
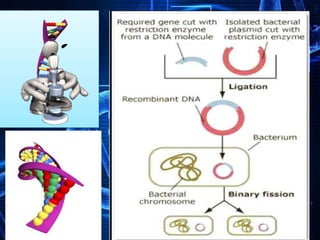
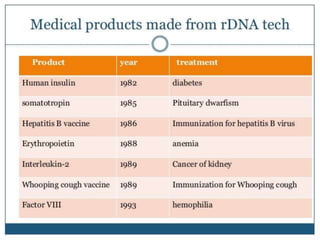
Recombinant DNA technology involves splicing DNA molecules from different sources to create new genetic combinations useful in various fields such as medicine, agriculture, and industry. Key techniques include isolating desired DNA, inserting it into vectors, and introducing recombinant DNA into host cells, with various tools like restriction enzymes and ligases facilitating the process. The technology offers significant advantages, such as producing insulin and developing genetically modified crops, contributing to advancements in health and agriculture.