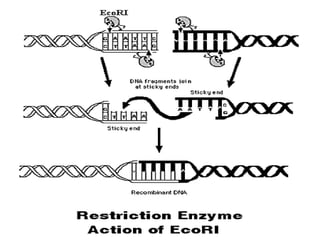
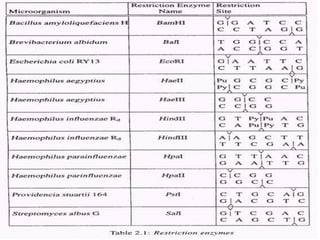
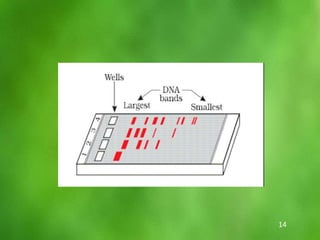
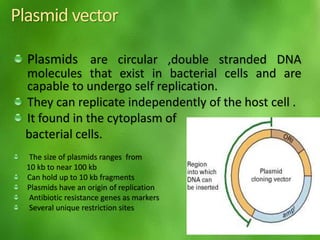
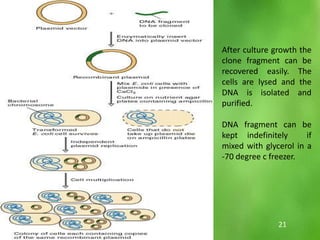
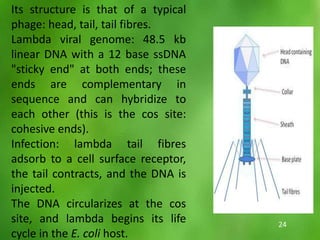
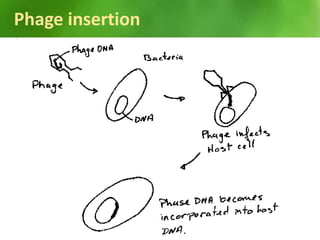
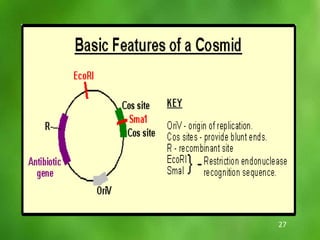
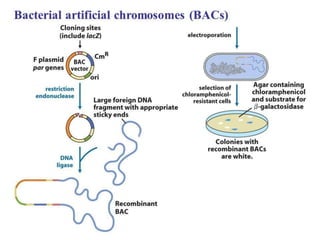
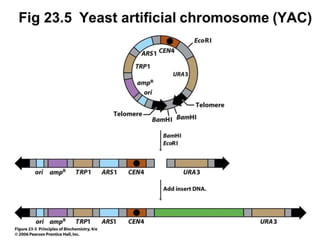
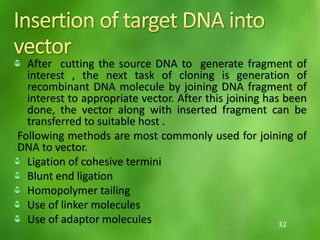
This document discusses rDNA technology and methods for gene cloning. It describes how recombinant DNA is formed by combining DNA sequences as desired. Gene cloning involves isolating a specific DNA fragment from an organism and introducing it into a plasmid vector that can replicate in a host cell, producing multiple copies of the fragment. Various methods are described for isolating DNA fragments, including mechanical shearing, restriction enzyme digestion, and reverse transcriptase. Different types of vectors like plasmids, bacteriophages, cosmids, BACs, and YACs that can be used are also summarized.