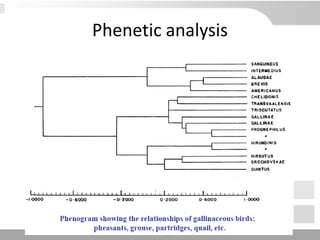
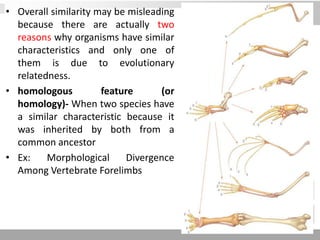
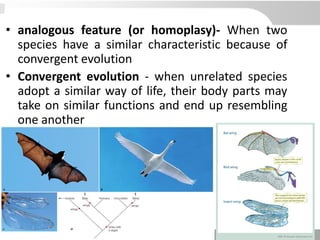
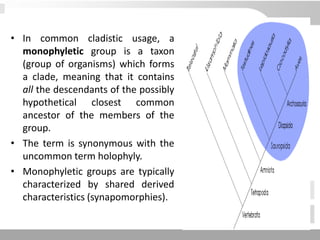
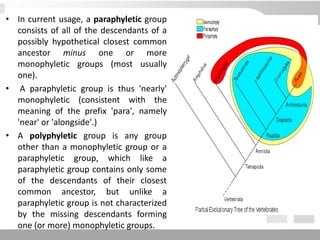
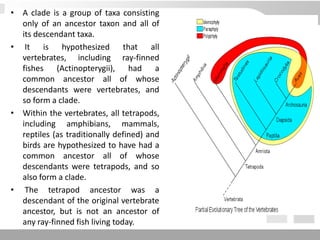
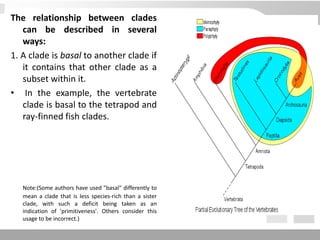
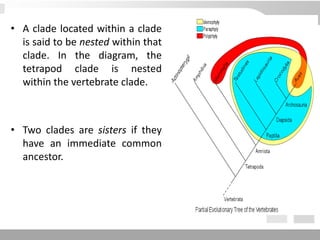
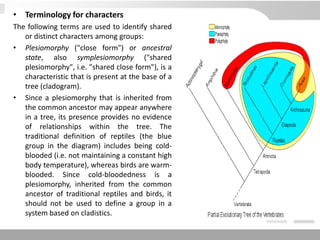
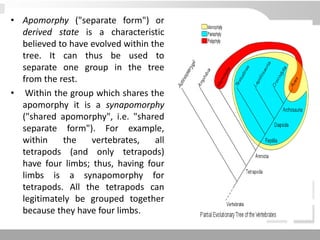
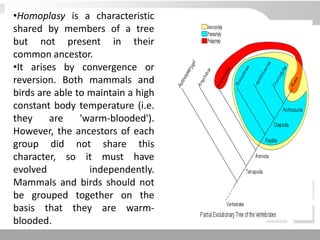
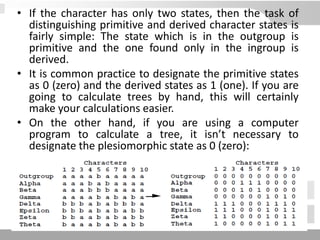
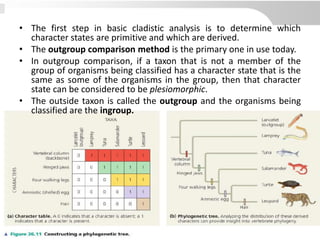
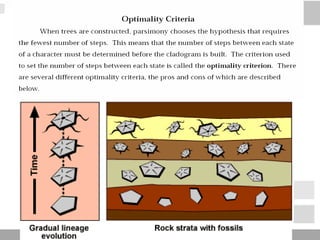
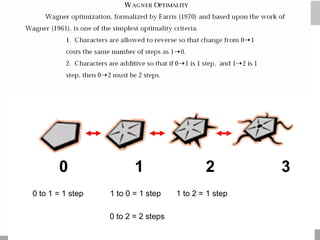
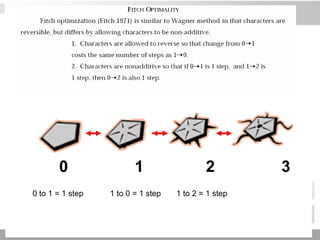
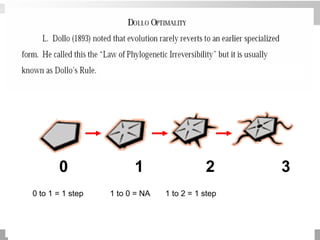
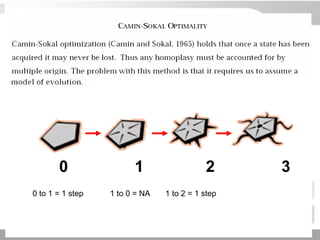
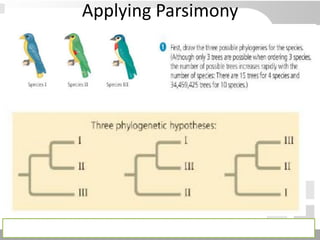
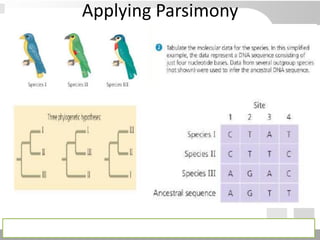
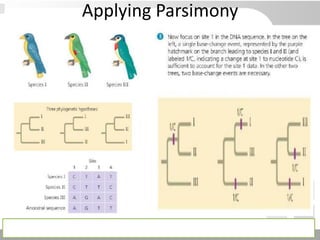
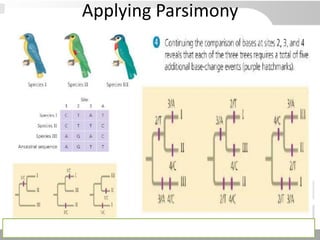
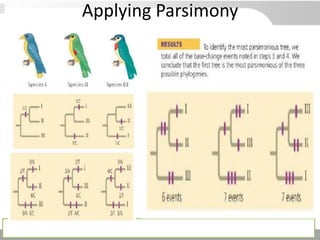
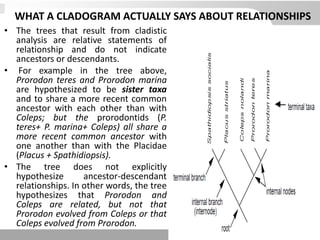
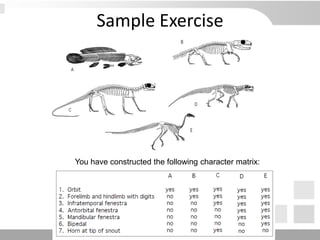
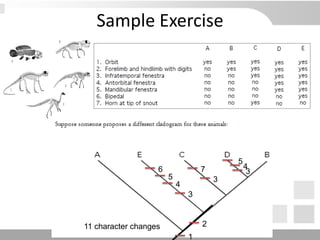
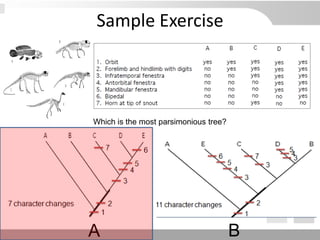
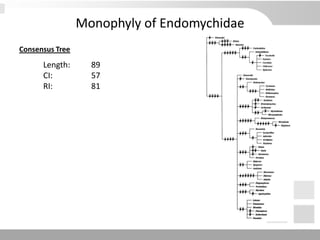
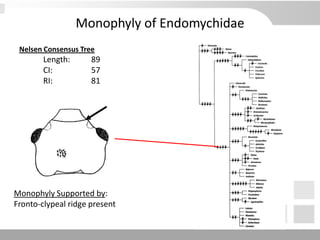
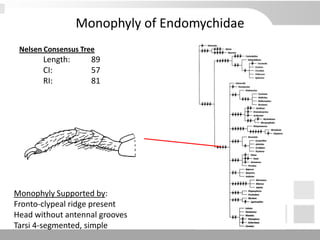
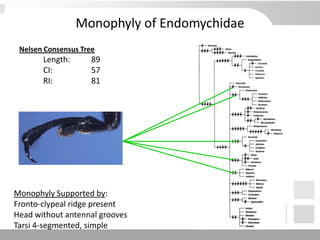
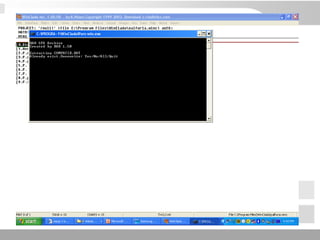
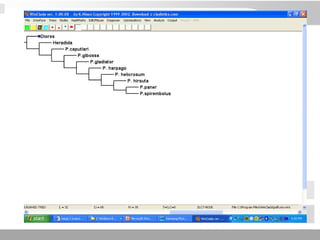
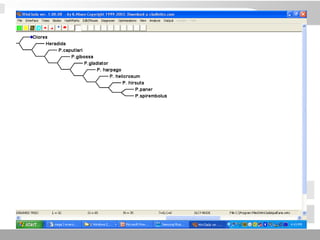
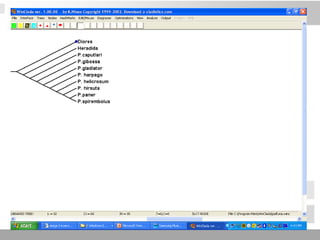
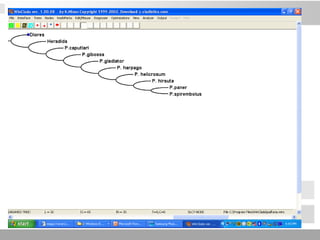
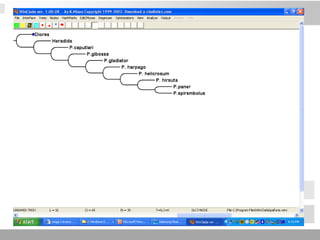
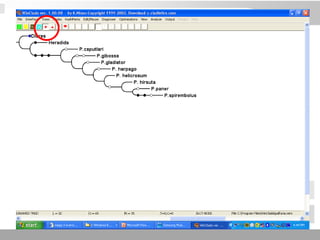
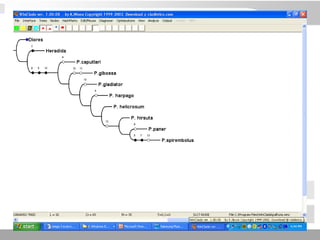
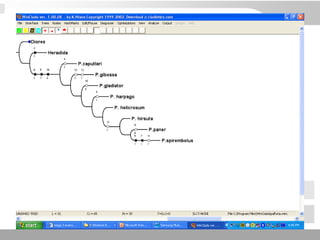
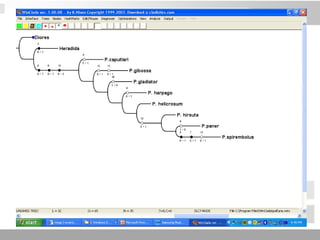
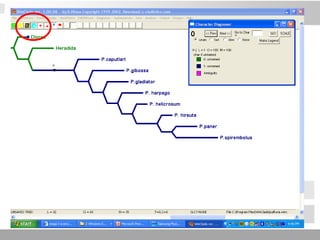
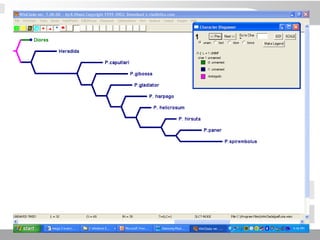
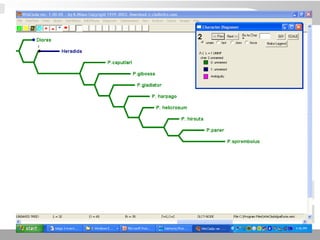
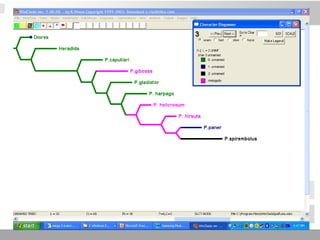
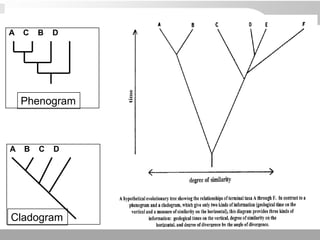
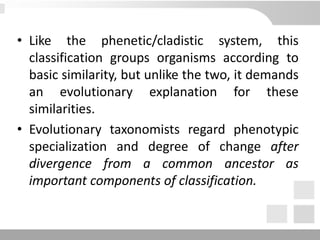
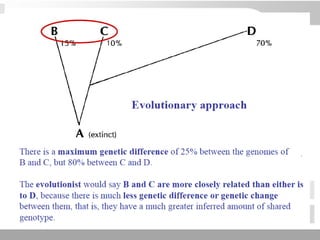
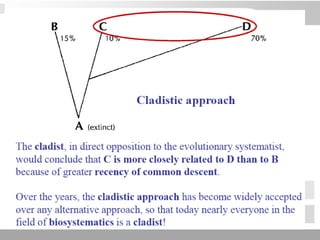
The document discusses three schools of systematics: phenetics, cladistics, and evolutionary classification, outlining their approaches to classifying organisms based on similarities and evolutionary relationships. It explains concepts such as homologous and analogous features, monophyletic and paraphyletic groups, and the importance of derived characteristics in understanding phylogenetic relationships. Additionally, it touches on the methodologies used in cladistics, including outgroup comparison and maximum parsimony, while highlighting the challenges in traditional taxonomic classifications.