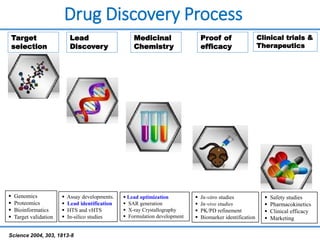
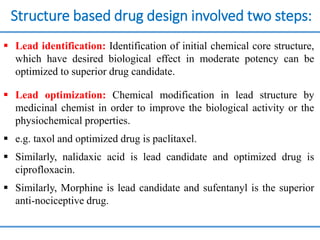
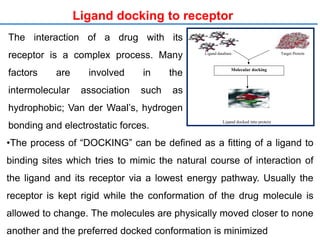
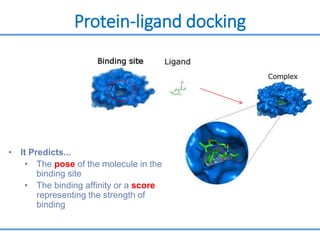
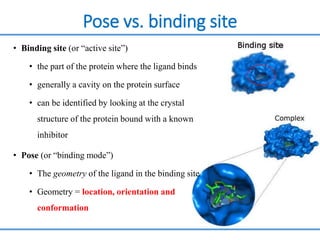
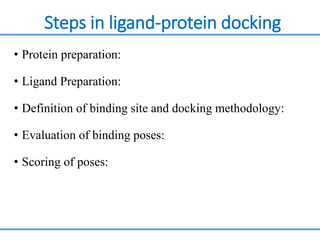
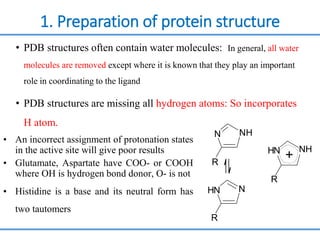
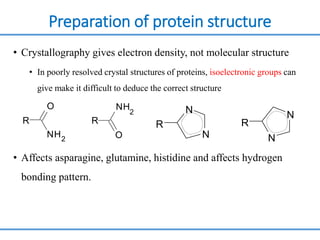
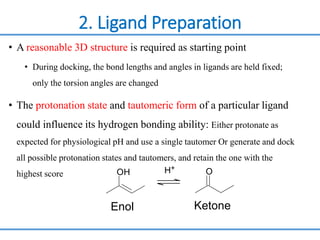
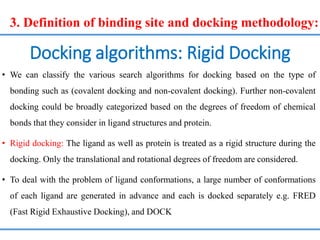
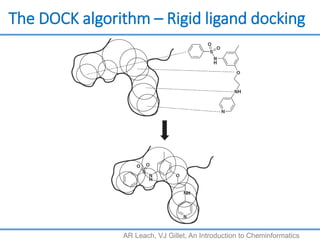
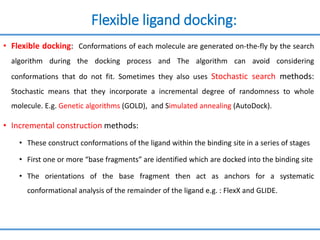
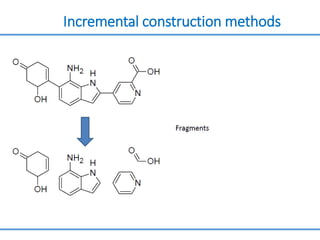
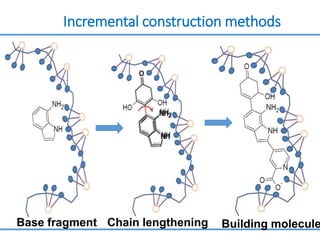
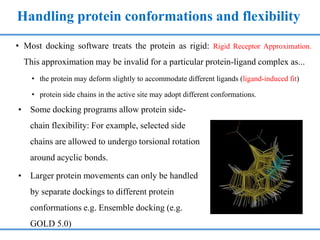
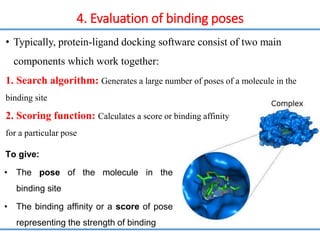
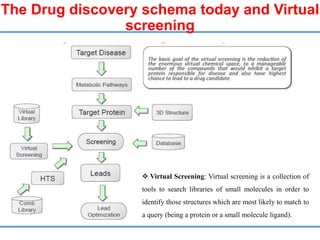
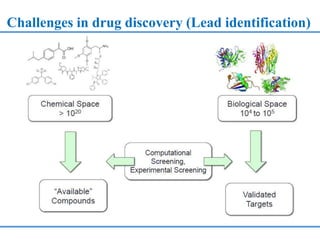
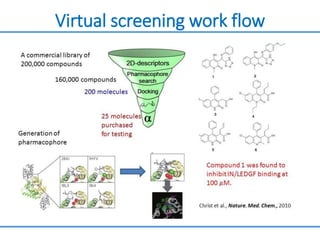
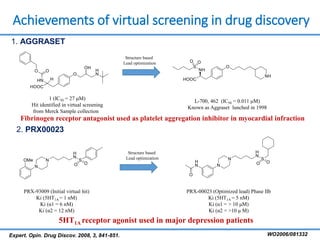
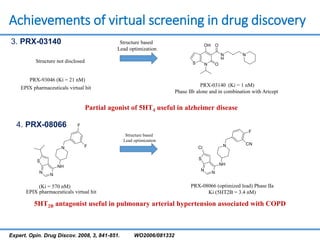
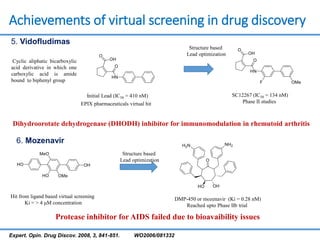
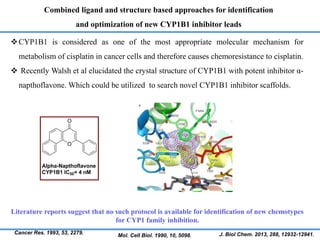
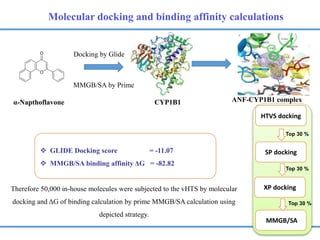
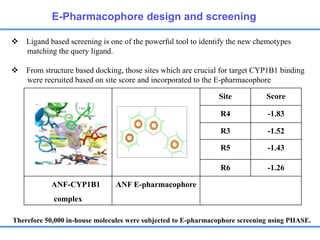
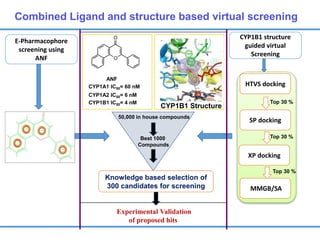
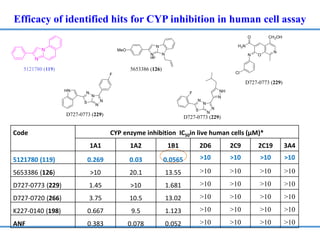
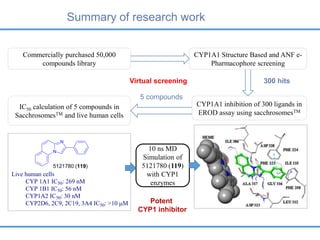
Structure based drug design involves identifying lead compounds through virtual screening and then optimizing these leads through medicinal chemistry. Key steps include target selection, virtual screening to identify initial hits, lead optimization through structure-activity relationship studies and molecular docking to refine binding, and progression through preclinical and clinical testing. Several successful drugs have been developed using this approach, starting from virtual screening hits that were subsequently optimized into drug candidates.