The document summarizes research on generating high-quality human reference genomes using PromethION nanopore sequencing. Key points:
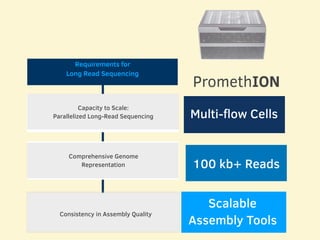
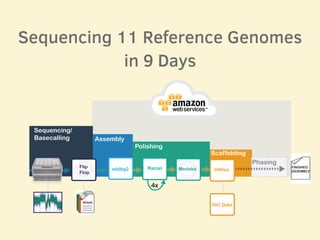
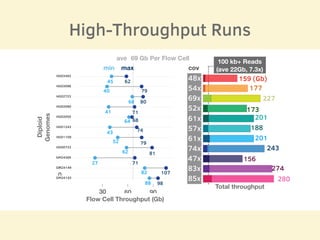
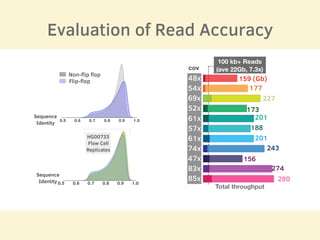
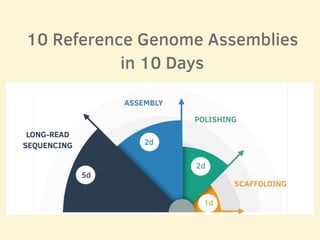
- 11 human reference genomes were sequenced in 9 days using PromethION nanopore sequencing and assembly tools, achieving finished assemblies.
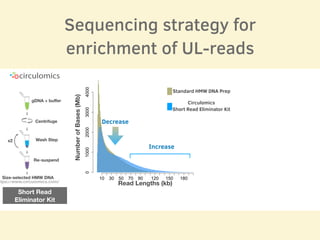
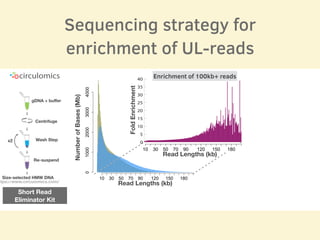
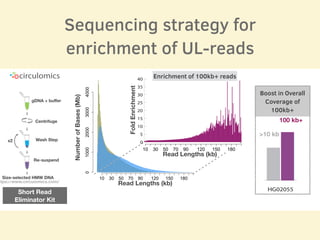
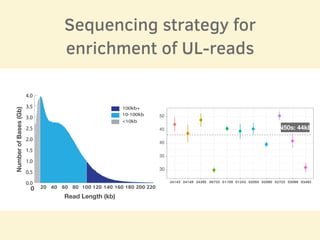
- The sequencing strategy included enriching for ultra-long reads over 100kb using a short read eliminator kit to boost overall coverage of long reads.
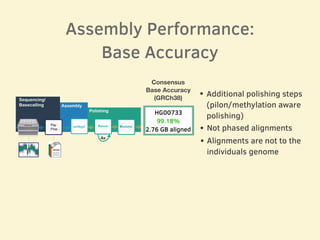
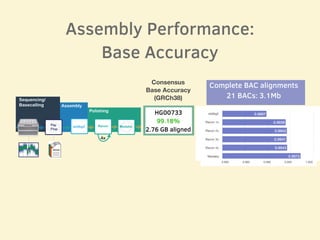
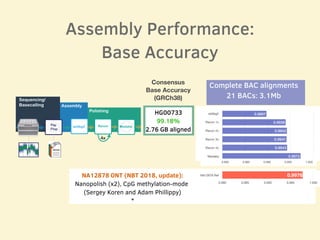
- Evaluation of one genome assembly showed over 99% consensus base accuracy when aligned to the human reference genome and over 99.76% accuracy for alignments of complete BAC sequences.
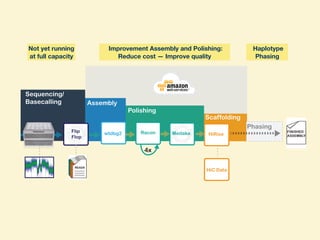
- The research aims to further improve assembly quality and reduce costs while increasing throughput using PromethION sequencing and optimized assembly tools