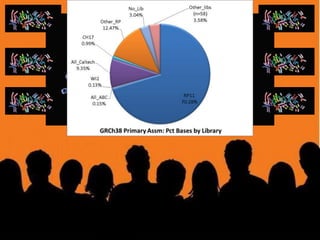
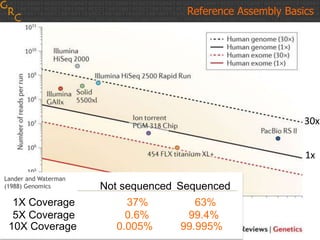
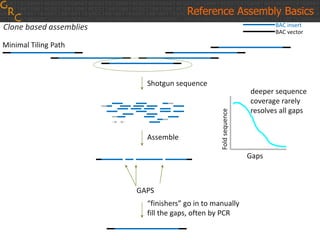
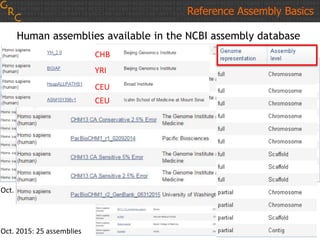
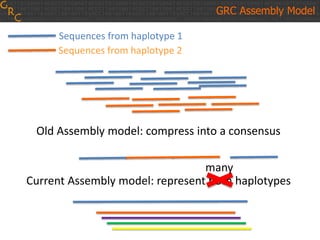
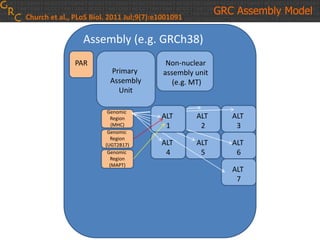
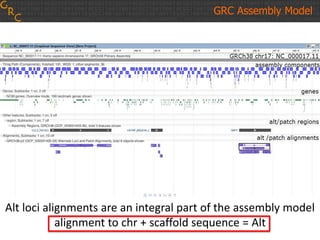
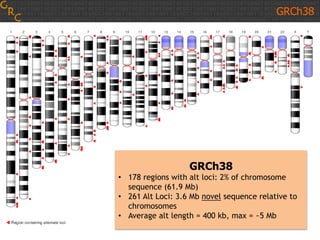
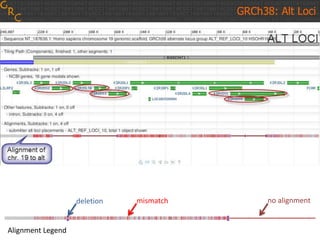
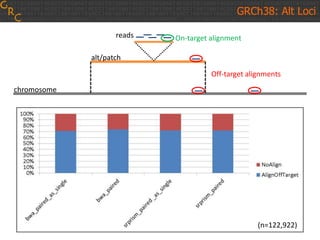
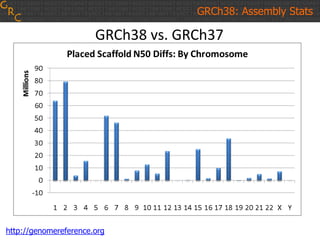
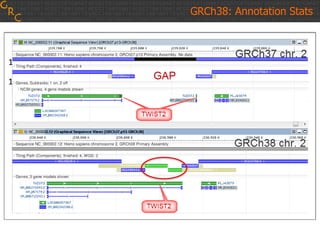
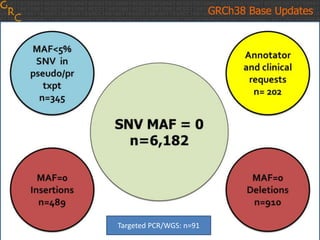
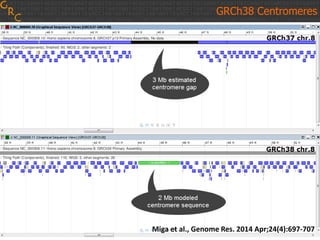
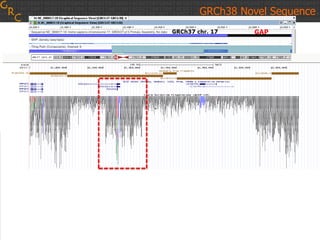
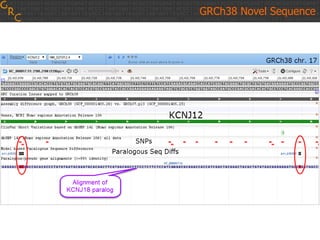
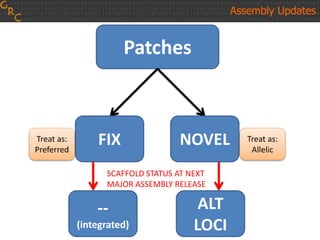
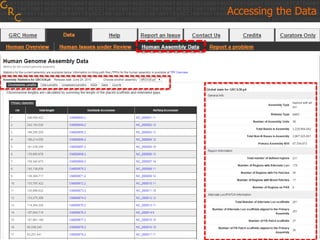
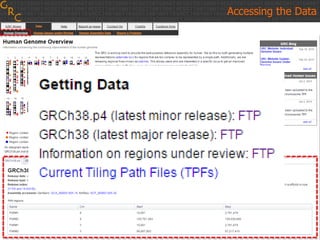
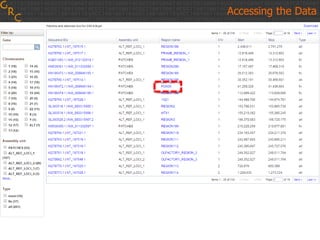
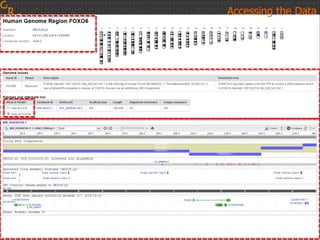
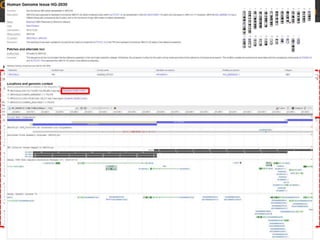
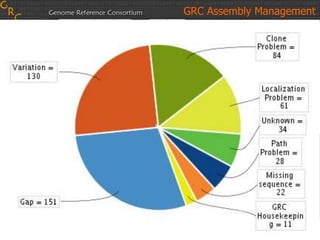
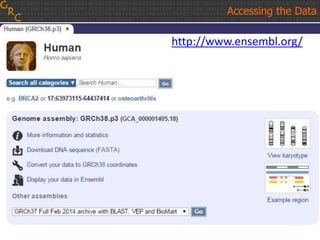
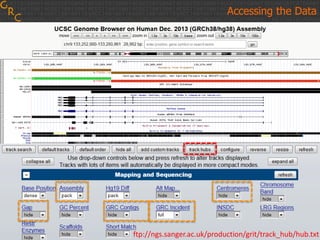
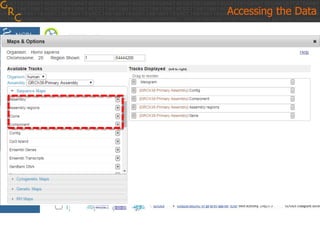
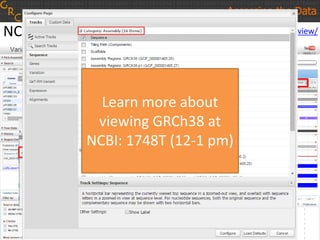
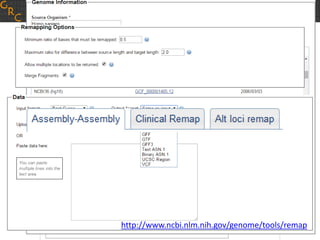
The document discusses the human reference genome assembly GRCh38. It provides information on assembly basics, the assembly model used, updates made in GRCh38 compared to the previous version, and how to access and utilize the sequence and annotation data. Key points include that GRCh38 represents 178 genomic regions with alternative loci sequences, contains over 400kb of novel sequence from targeted updates, and is collaboratively maintained to integrate new data.