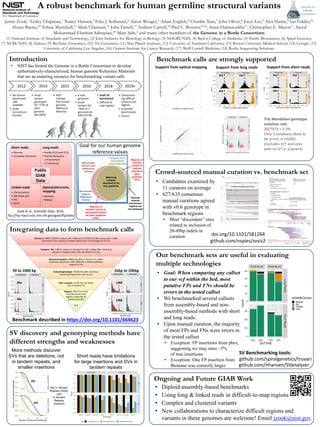
1) Discovery: Over 1 million structural variant calls were discovered across 30 sequence-resolved callsets from 4 technologies for an AJ Trio. After clustering, over 128,000 sequence-resolved calls remained.
2) Discovery Support: Over 30,000 structural variants had support from 2+ technologies or 5+ callers in the trio.
3) Evaluate/genotype: Nearly 20,000 structural variants had a consensus variant genotype predicted for the son from analyzing the trio.