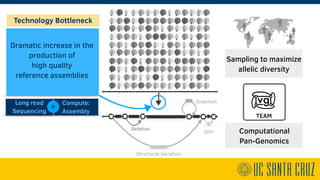
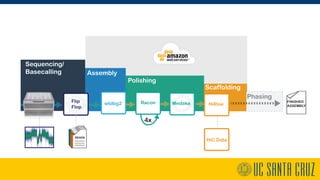
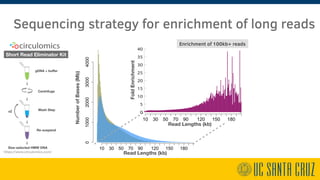
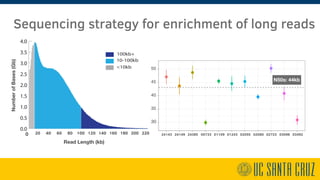
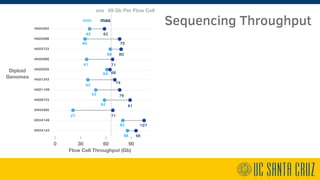
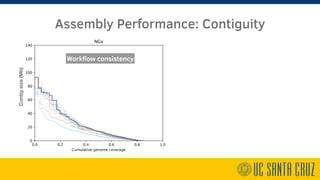
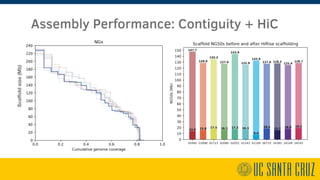
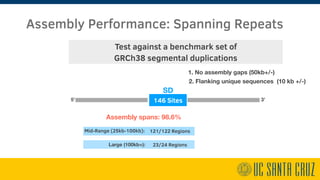
The document describes using PromethION nanopore sequencing to generate high-quality human reference genomes. 11 reference genomes were sequenced in 9 days using PromethION, achieving high consensus accuracy (>99%) and continuity. The approach leverages long reads for assembly followed by polishing and scaffolding. This high-throughput and accurate method can generate reference genomes at an estimated cost of $10,000 per genome.