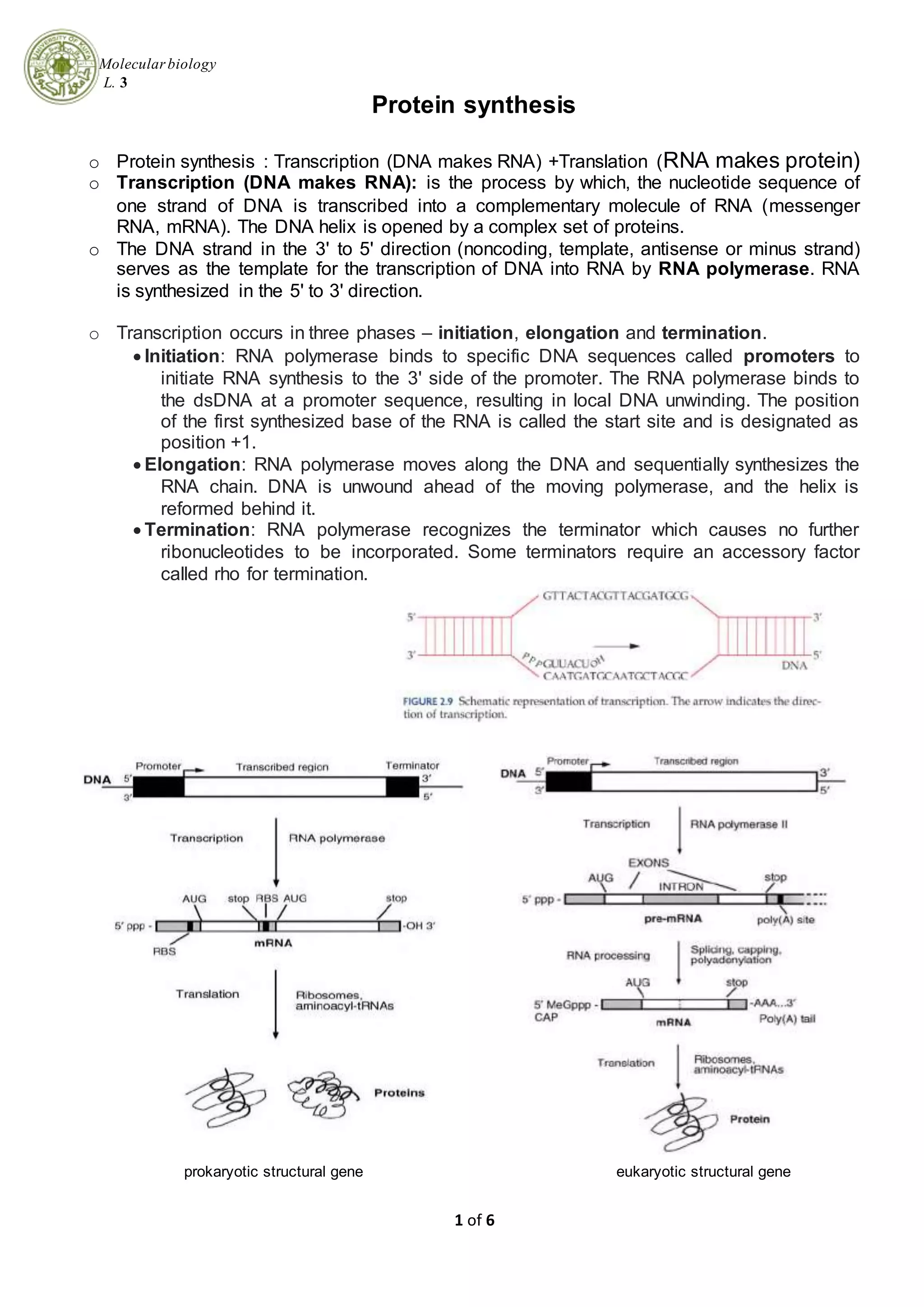
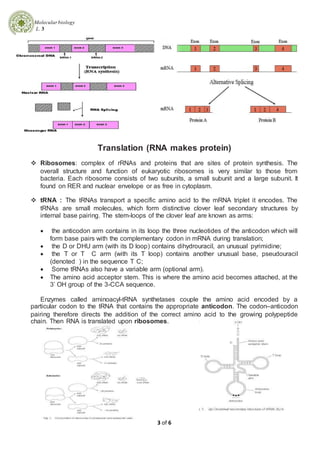
1. Transcription is the process by which DNA is copied into messenger RNA (mRNA) by RNA polymerase. This involves three phases - initiation, elongation, and termination.
2. Eukaryotic transcription is more complex than prokaryotic transcription due to multiple RNA polymerases, nucleosomes, separation of transcription and translation, and intron-exon structure of genes.
3. Following transcription, eukaryotic mRNA undergoes processing including capping, polyadenylation, and splicing before being translated into protein by ribosomes.