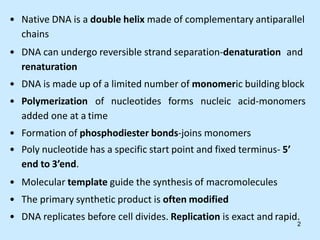
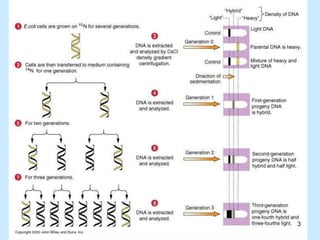
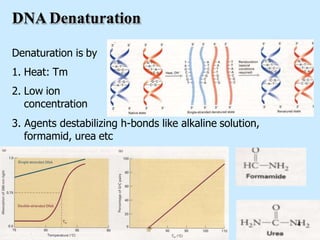
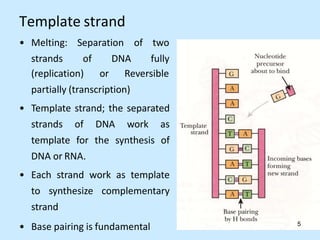
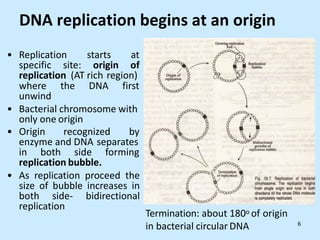
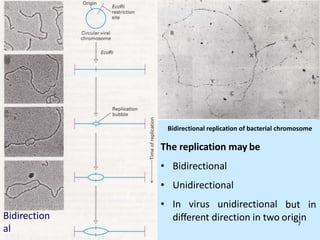
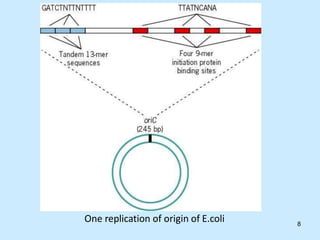
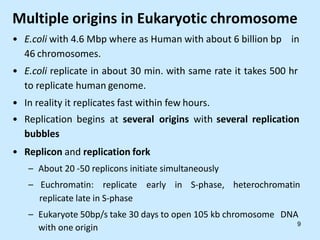
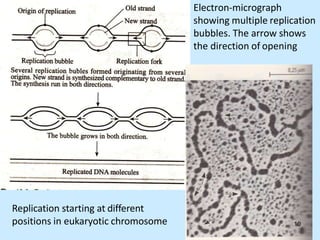
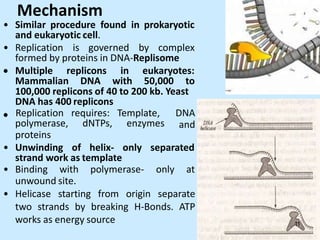
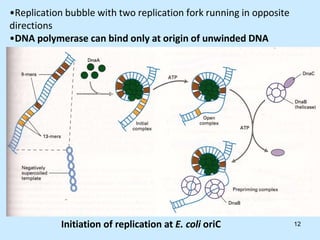
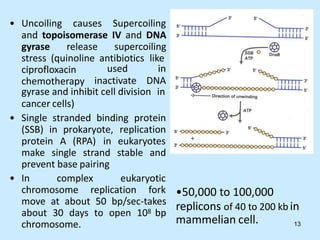
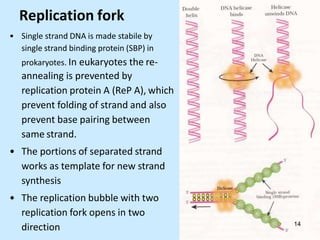
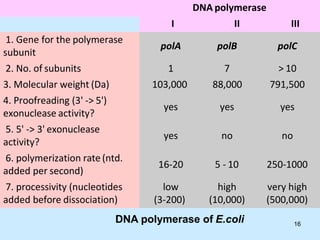
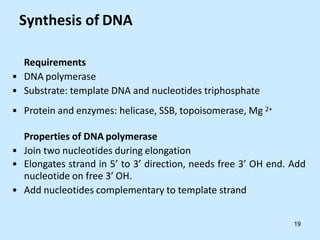
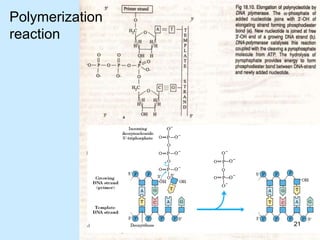
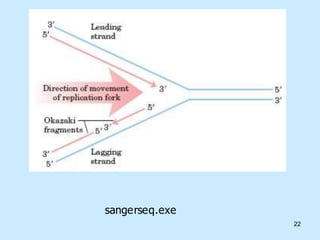
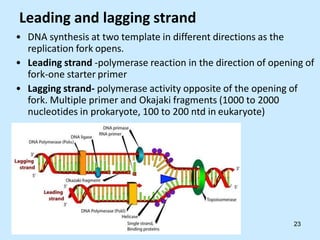
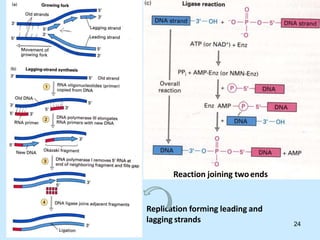
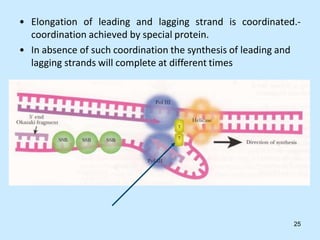
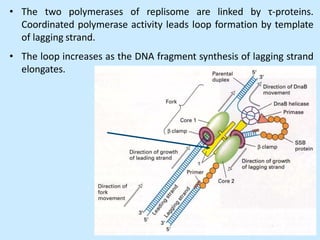
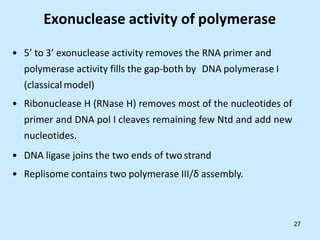
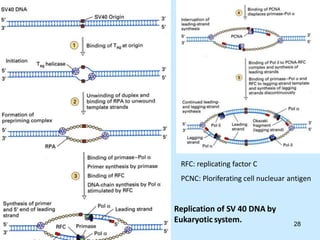
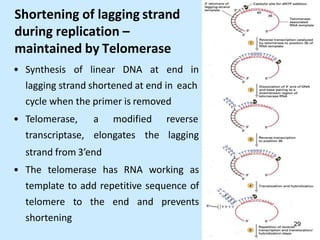
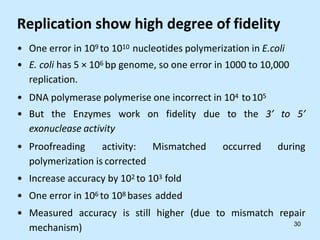
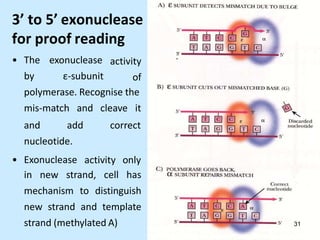
DNA replication is semi-conservative and begins at origins of replication. In eukaryotes, replication initiates from multiple origins and proceeds bidirectionally. The double helix separates into single strands through the action of helicase. DNA polymerase adds nucleotides to the 3' end of the growing strand based on complementary base pairing. Leading and lagging strands are synthesized differently due to their direction of synthesis relative to the replication fork. Fidelity is ensured by proofreading exonuclease activity and mismatch repair systems.