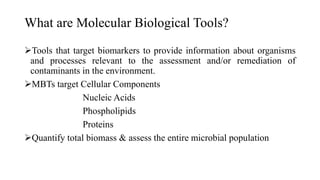
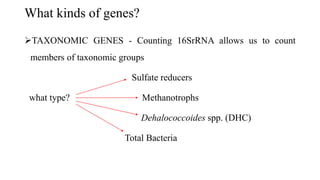
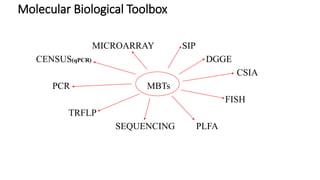
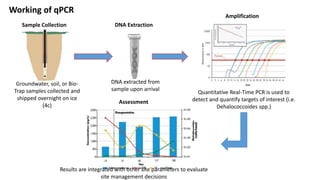
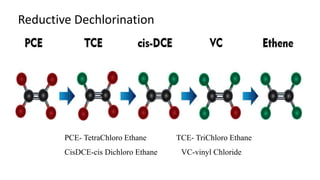
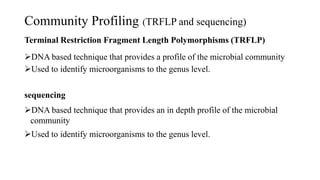
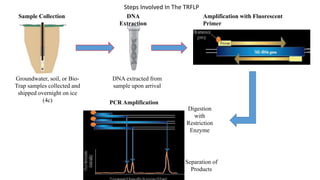
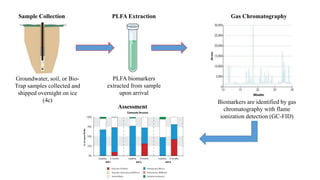
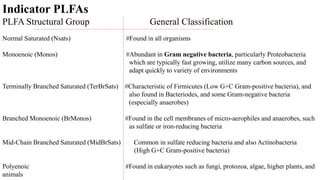
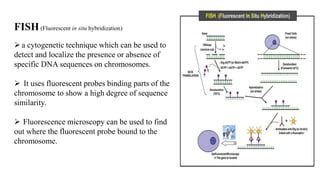
The document discusses molecular biological tools (MBTs) used for environmental management, which include techniques like quantitative PCR, PLFA analysis, and microarray-based assessments, focusing on the quantification of microorganisms involved in biodegradation processes. It highlights advantages over traditional microbiology, such as improved representation of in-situ communities and the ability to track specific functional genes. The document also outlines various sampling methods and applications for evaluating and managing environmental contaminants.