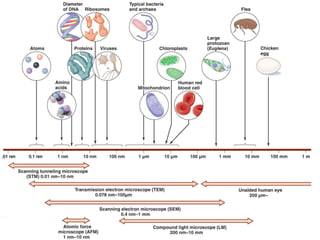
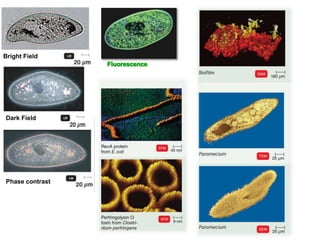
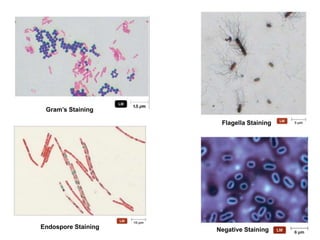
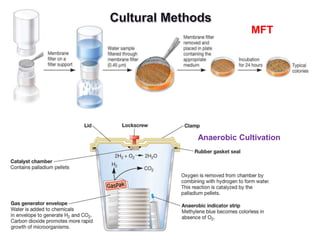
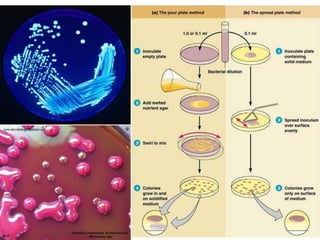
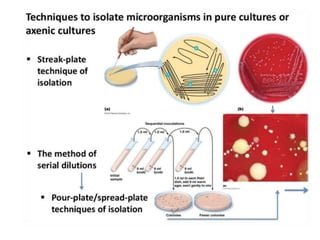
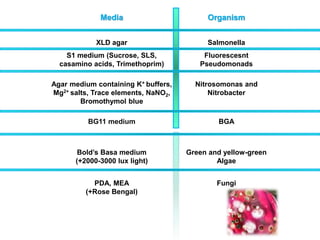
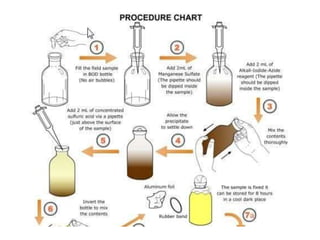
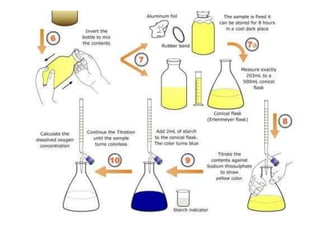
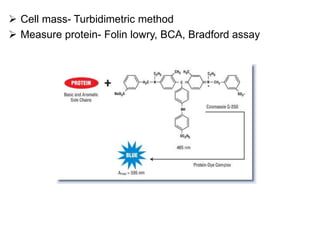
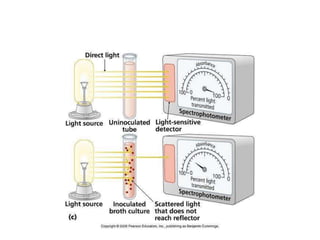
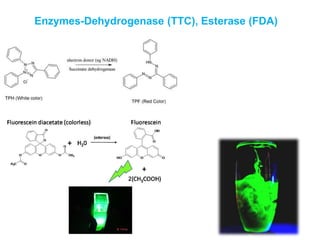
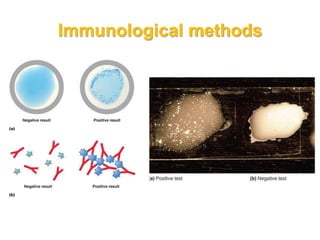
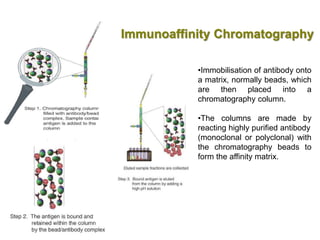
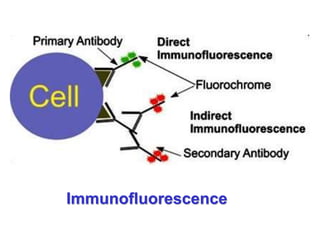
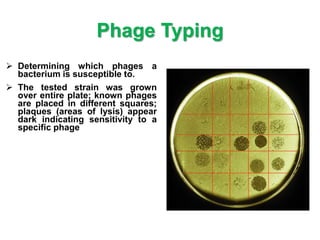
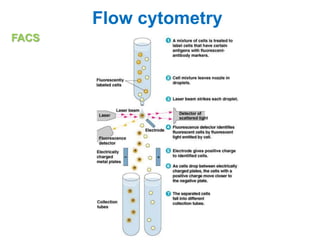
This document discusses various microbiological techniques used to study microorganisms, including:
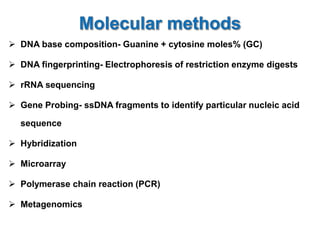
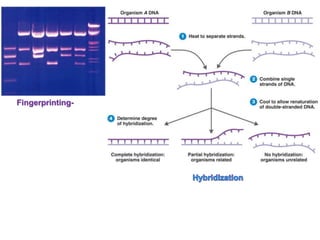
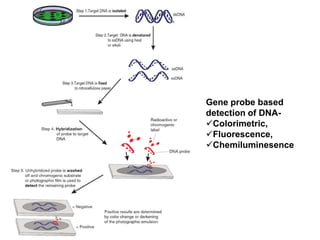
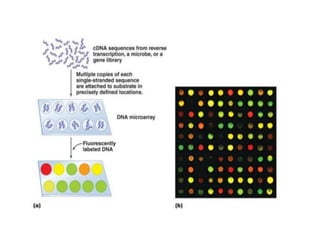
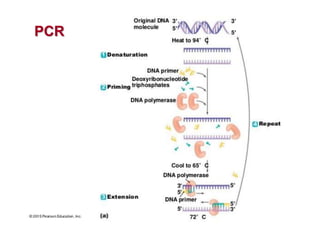
1. Microscopic, cultural, physiological, immunological, and molecular methods. Specific techniques mentioned are Gram staining, growth media selection, enzyme activity assays, immunoassays, DNA fingerprinting, gene probes, microarrays, PCR, and metagenomics.
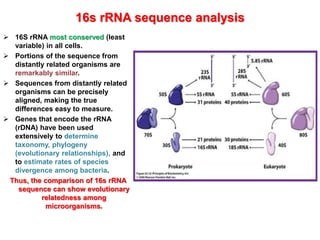
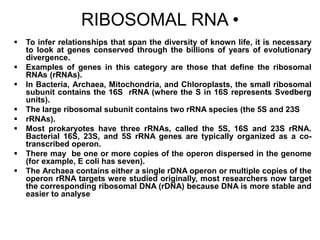
2. 16S rRNA gene sequencing is described as the most widely used molecular technique for bacterial identification and phylogenetic analysis due to the conserved nature of the 16S rRNA gene.
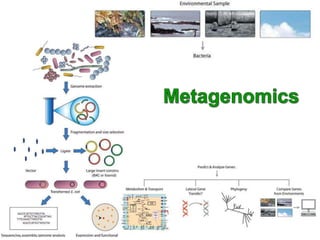
3. Metagenomics provides information on the collective genomes of microorganisms in an environmental sample to study microbial diversity and ecology.