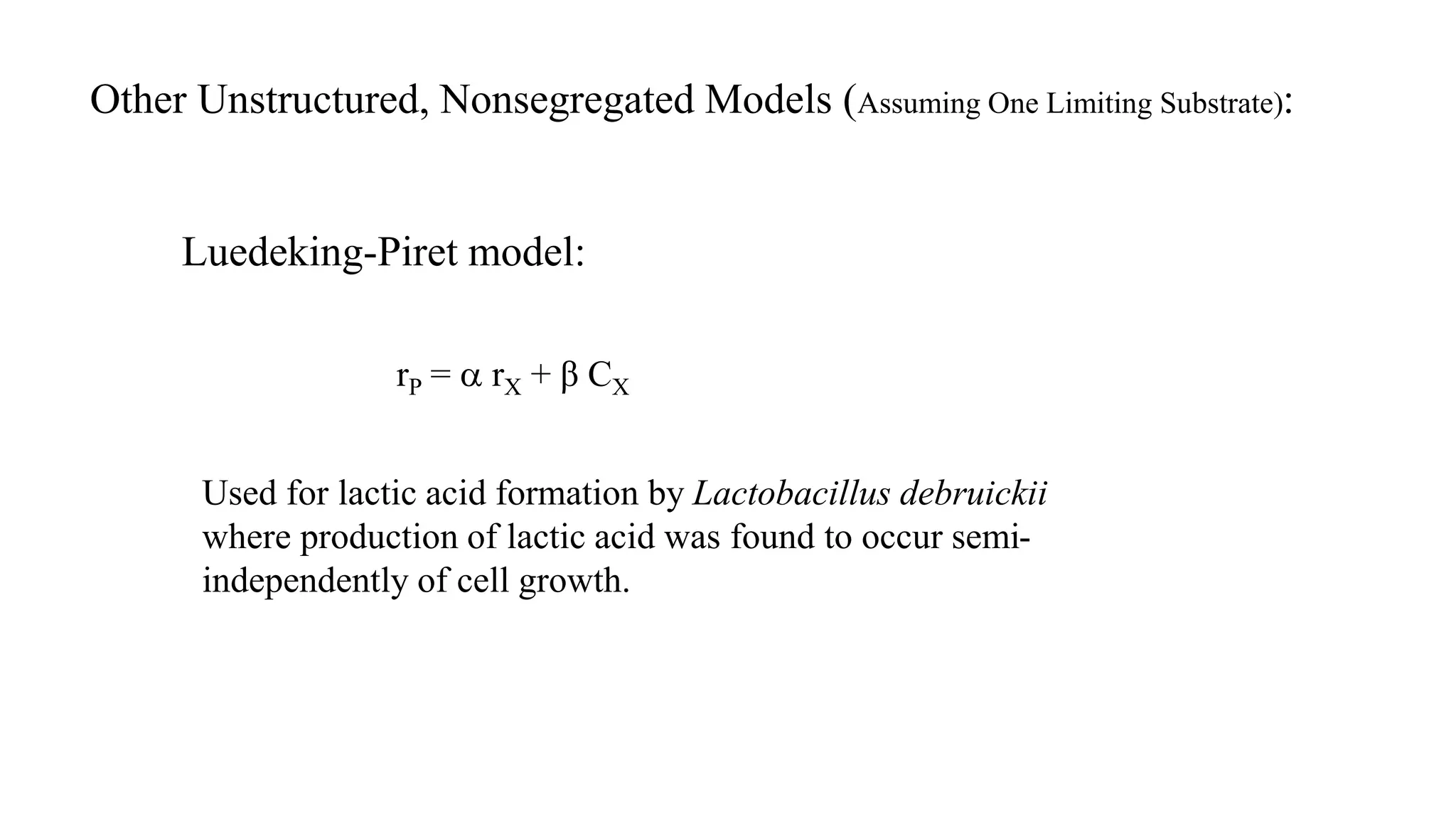
This document discusses unstructured, non-segregated models used to model population-level cell growth and metabolism. The Monod model is the most commonly used model that describes growth rate as a function of substrate concentration. Several variants of the Monod model are presented to account for substrate inhibition, product inhibition, and toxic compound inhibition. Other unstructured models discussed include the Blackman, Tessier, Moser, Contois, and Luedeking-Piret models. The assumptions and limitations of unstructured, non-segregated models are also summarized.









![Other unstructured, nonsegregated models (assuming one limiting substrate):
Blackman equation: μ = μm if CS ≥ 2KS
μ =
μm CS
2 KS
if CS < 2KS
Tessier equation: μ = μm [1 - exp(-KCS)]
Moser equation: μ =
μm CS
n
KS + CS
n
Contois equation: μ =
μm CS
KSX CX + CS](https://image.slidesharecdn.com/unstructuredmodelonpopulationlevel-150810183915-lva1-app6892/75/Unstructured-model-on-population-level-10-2048.jpg)

![Prof. R. Shanthini
Tessier equation:
0
0.2
0.4
0.6
0.8
1
0 2 4 6 8 10
Cs (g/L)
μ(perh)
μm = 0.9 per h
K = 0.7 g/L
μ = μm [1 - exp(-KCS)]](https://image.slidesharecdn.com/unstructuredmodelonpopulationlevel-150810183915-lva1-app6892/75/Unstructured-model-on-population-level-12-2048.jpg)