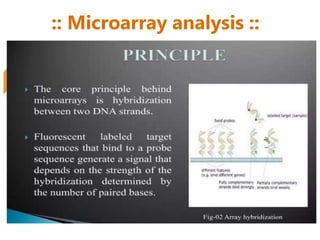
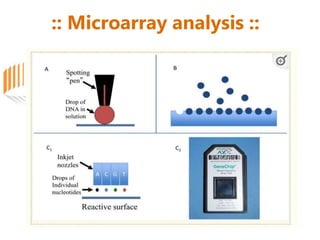
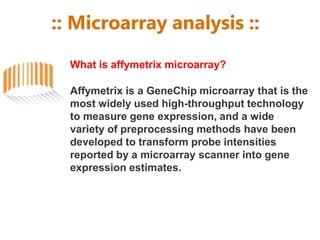
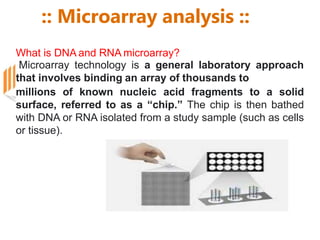
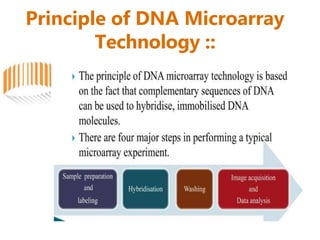
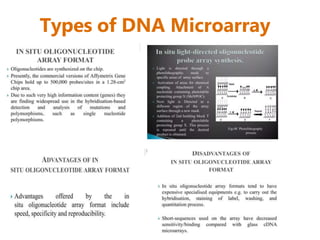
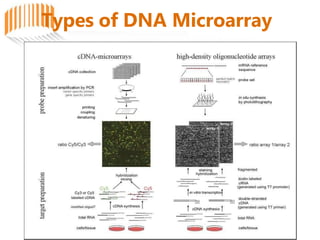
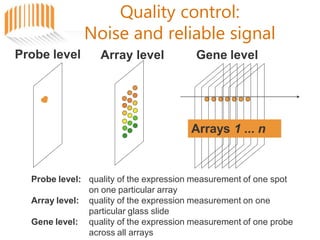
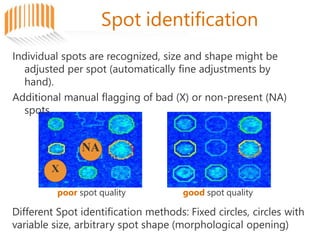
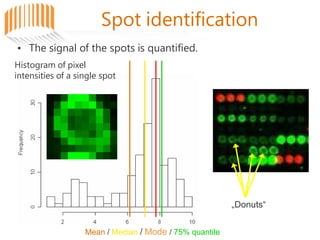
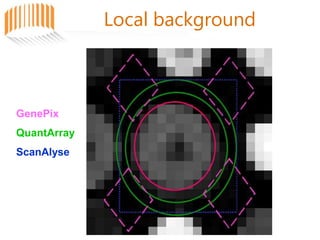
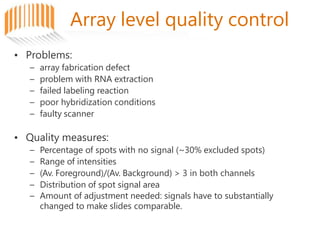
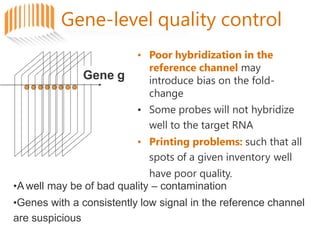
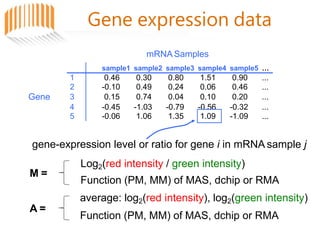
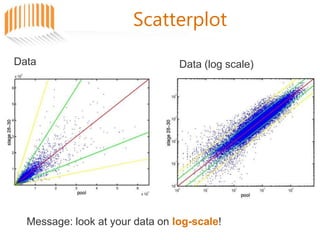
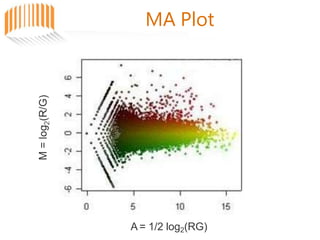
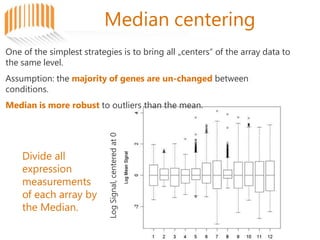
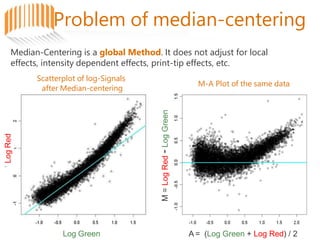
Microarrays allow researchers to analyze gene expression across thousands of genes simultaneously. There are three basic types of microarrays: spotted arrays where DNA is physically deposited on a slide; self-assembled arrays using beads containing oligos in pits on a surface; and in-situ synthesized arrays produced by inkjet oligo synthesis or photolithography. Quality control is important at multiple levels including the probe, array, and gene levels to identify unreliable measurements and ensure only high quality data is used in analysis.