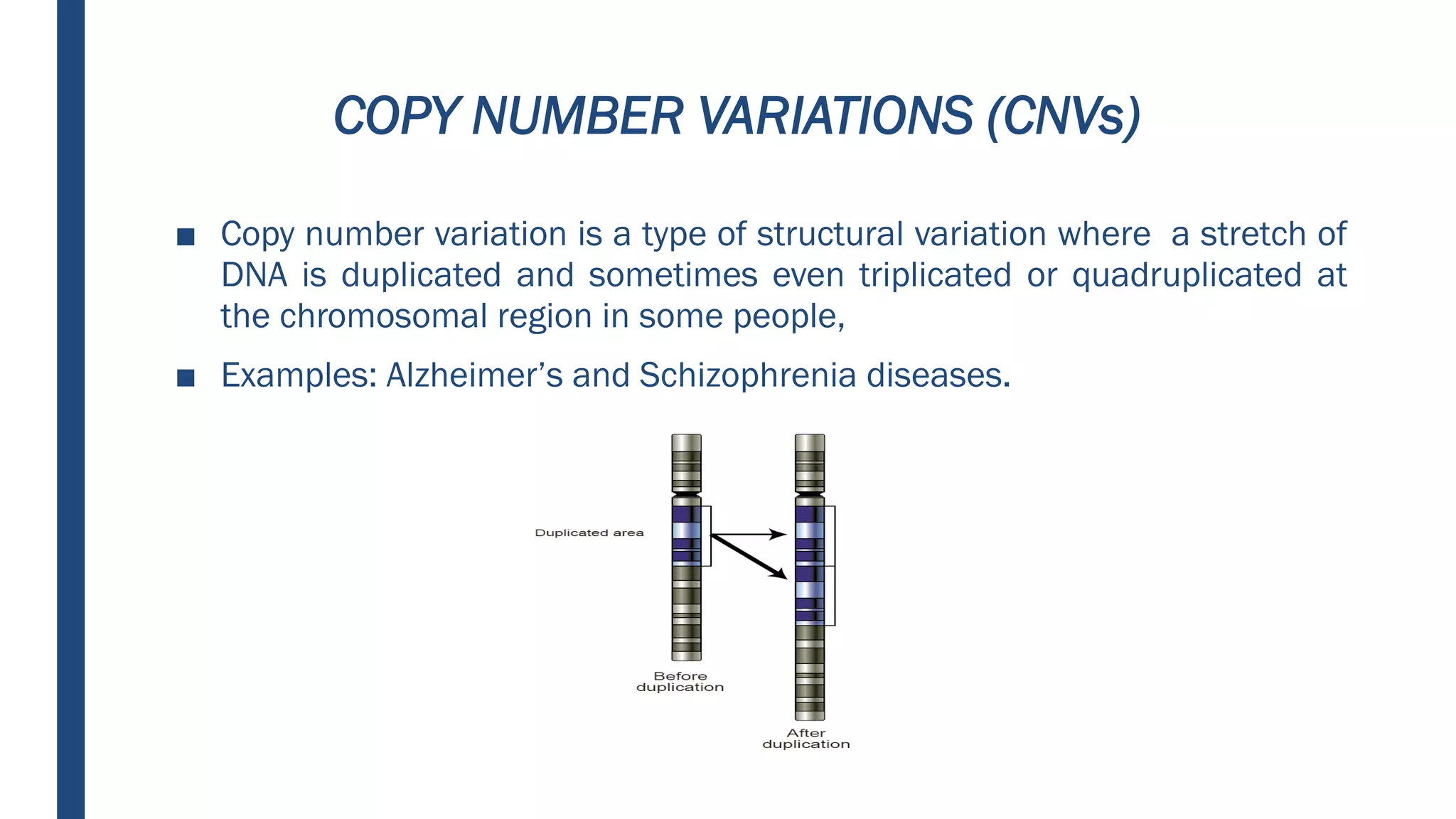
Human genetic variations include single nucleotide substitutions, tandem repeats, insertions and deletions, copy number variations, inversions, and translocations. These variations differ in DNA sequence and range in size from single nucleotides to megabases. The most common type is single nucleotide polymorphisms (SNPs), which represent differences in single nucleotides occurring about once every 1,000 nucleotides. SNPs can cause diseases like sickle cell anemia. Tandem repeats and copy number variations can also lead to diseases if DNA is misaligned during replication. Inversions and translocations occur when chromosomes break and rearrange, which can cause cancers, infertility, or Down syndrome. Variants in coding regions can be silent, nonsense, or missense mutations affecting protein function