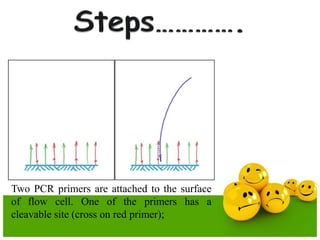
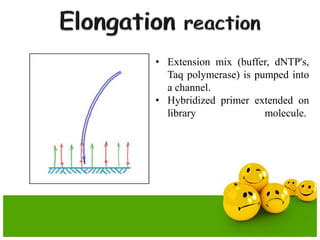
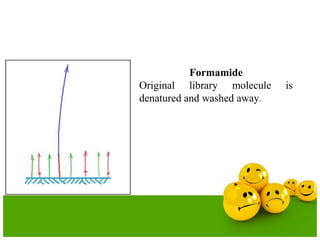
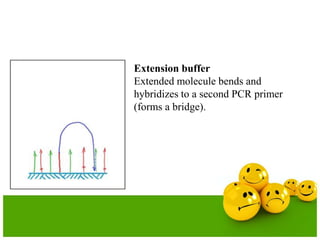
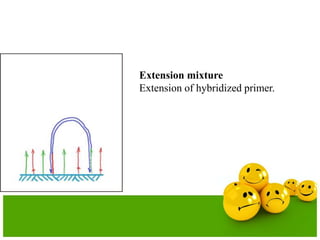
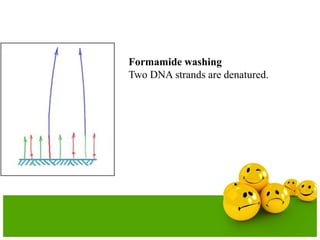
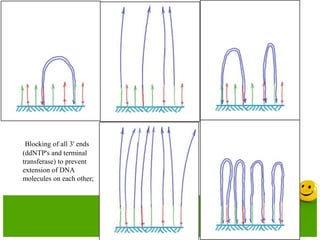
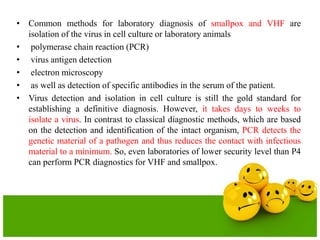
The document describes the process and components of emulsion PCR. Key points include:
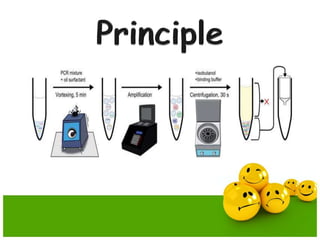
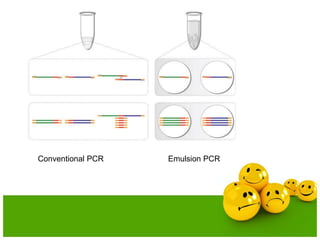
- Emulsion PCR is used to amplify DNA in microreactors formed from water-in-oil emulsions, allowing individual DNA fragments to be amplified clonally.
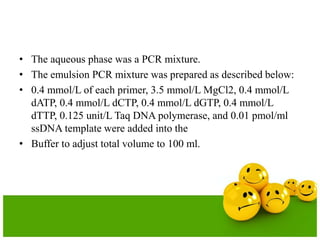
- The emulsion PCR mixture contains primers, DNA polymerase, nucleotides, template DNA, and is emulsified in an oil phase containing surfactants to form water-in-oil droplets.
- The emulsion undergoes PCR cycling to amplify the DNA fragments clonally within individual droplets. The emulsion is then broken and the amplified DNA fragments can be analyzed by gel electrophoresis or used for downstream applications like sequencing.