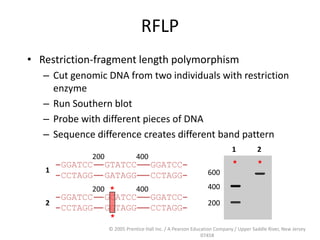
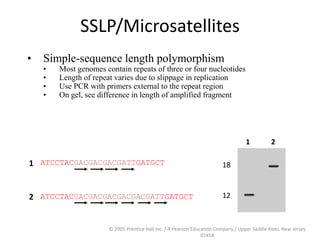
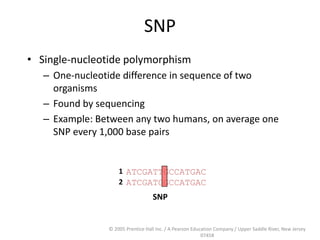
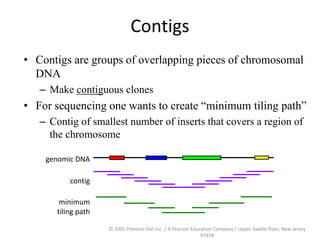
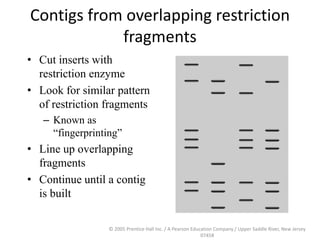
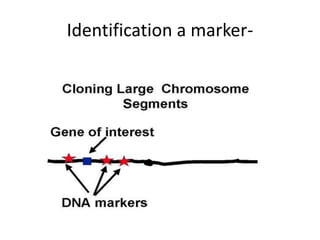
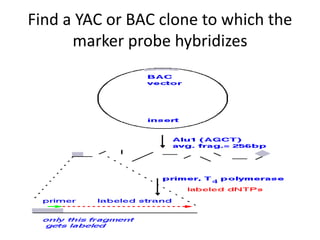
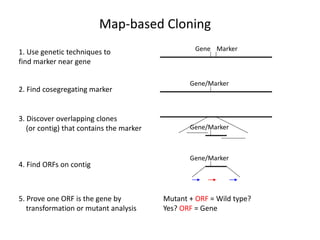
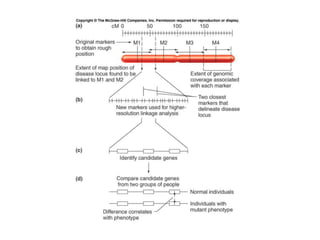
The document discusses map-based cloning of genomes, detailing its history, methods—including genetic and physical mapping—and practical applications. It outlines the steps involved in map-based cloning, such as identifying linked markers and characterizing candidate genes, while also highlighting genetic and physical markers used in the process. The conclusion emphasizes the technology's potential to clone necessary nucleotide sequences for various applications like genetic resistance and disease diagnosis.