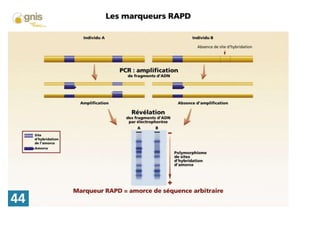
Random amplified polymorphic DNA (RAPD) is a type of PCR that uses short, arbitrary primers to randomly amplify DNA fragments. Several primers are used in PCR with genomic DNA to generate unique band patterns. RAPD does not require prior knowledge of DNA sequences and can detect mutations if they occur where the primer binds. However, it has lower resolution than targeted methods and results can be difficult to interpret. RAPD analyzes 100 to 3000 base pair fragments and computer programs can analyze the profiles.