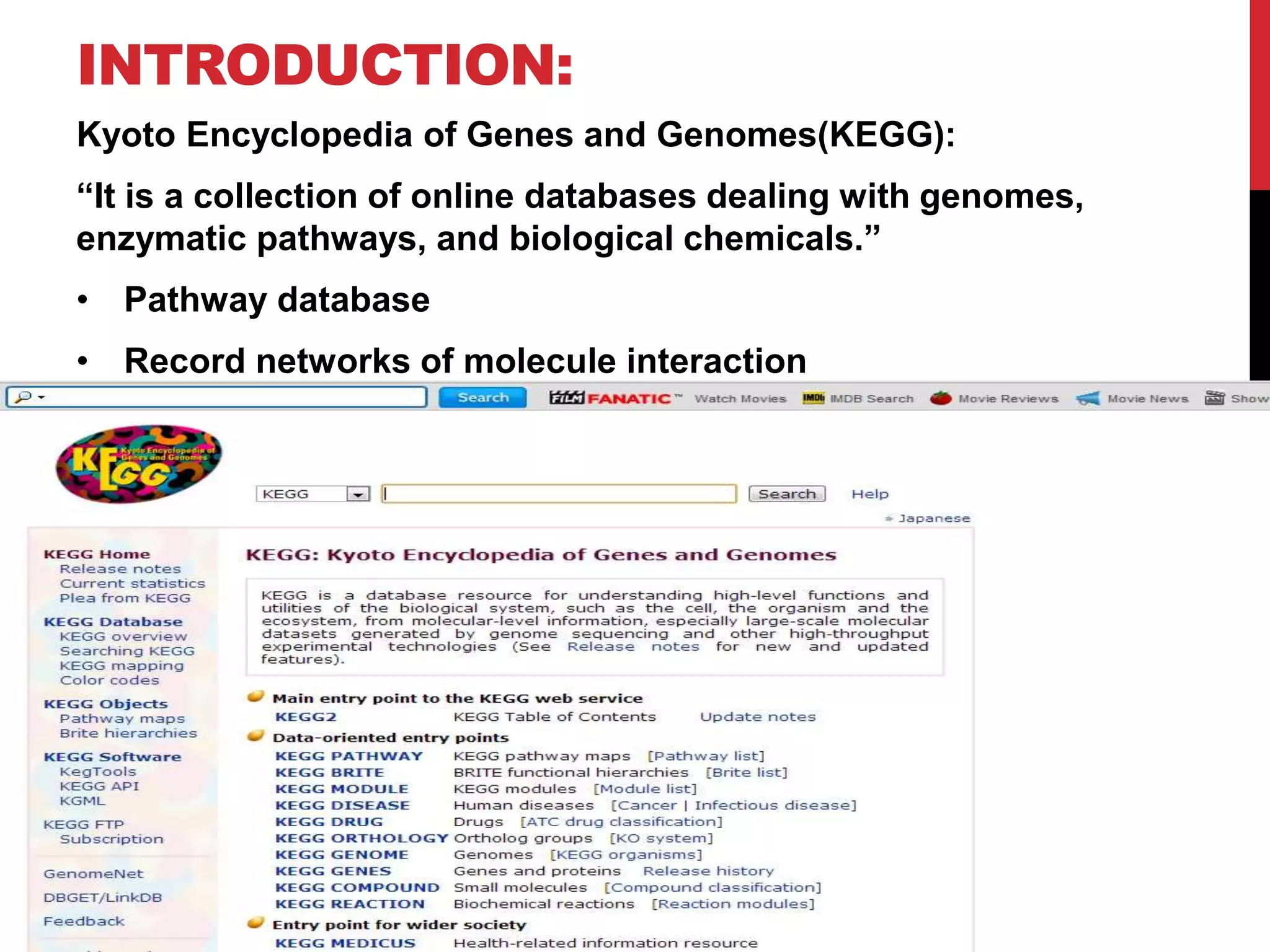
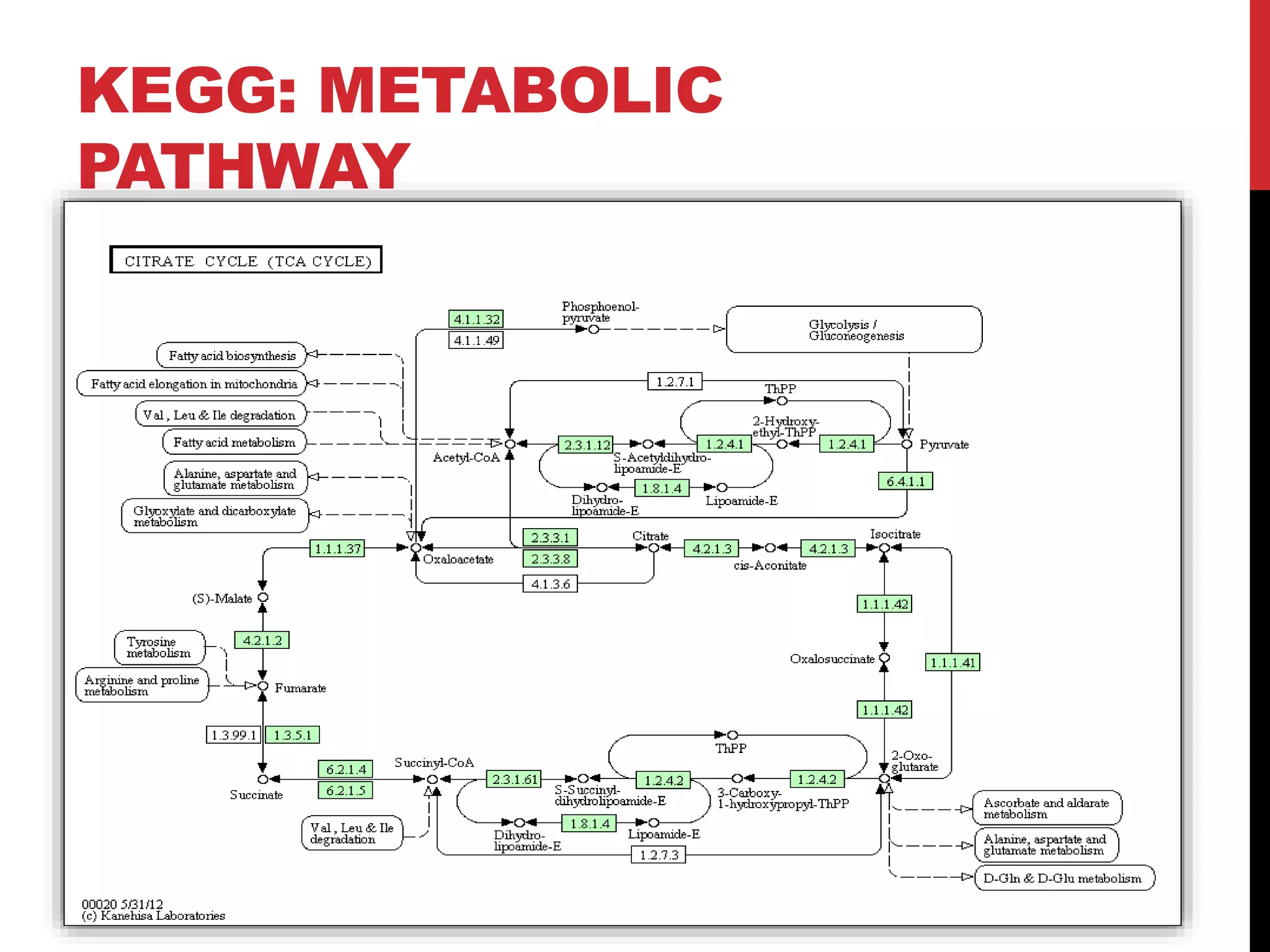
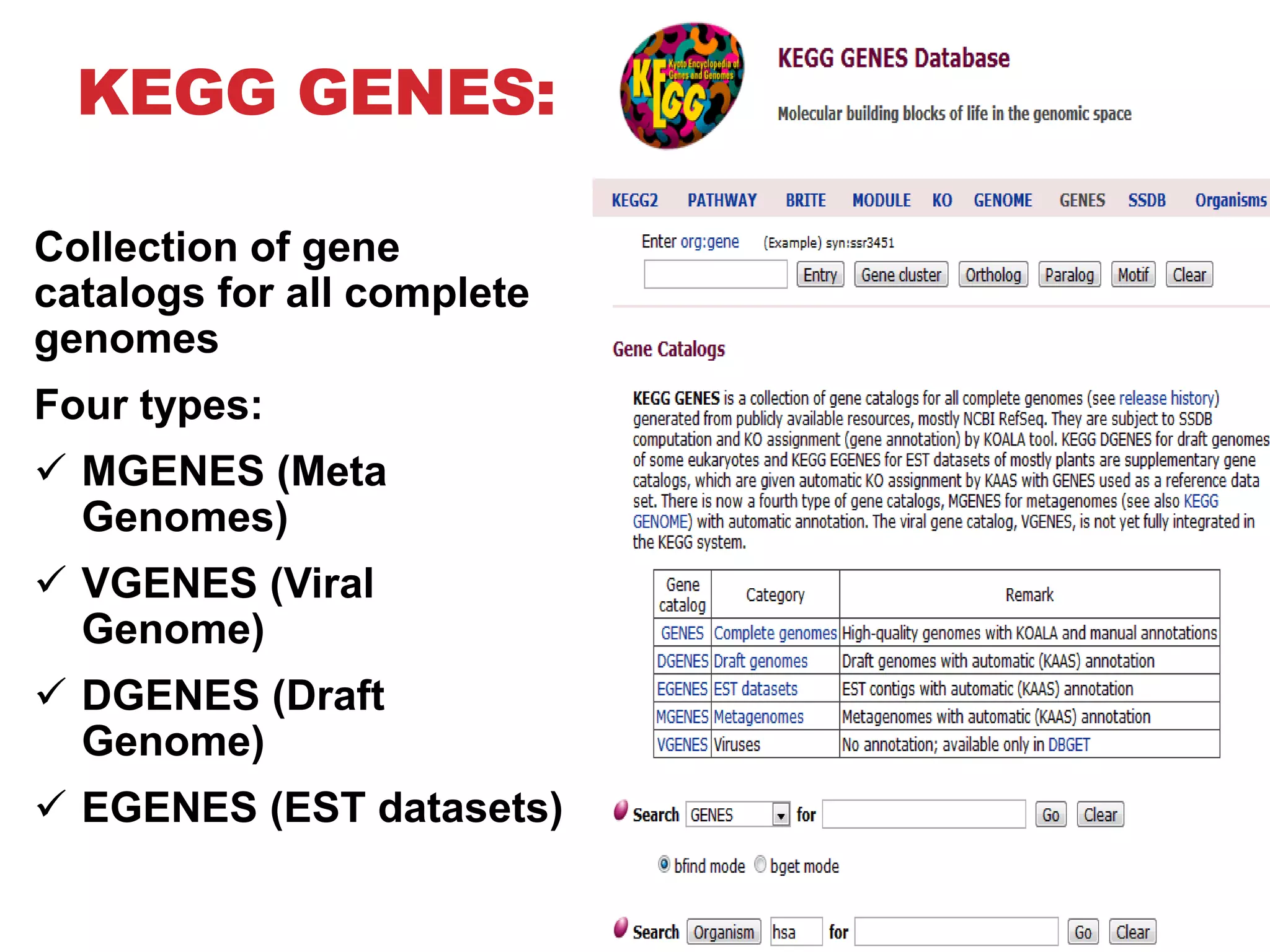
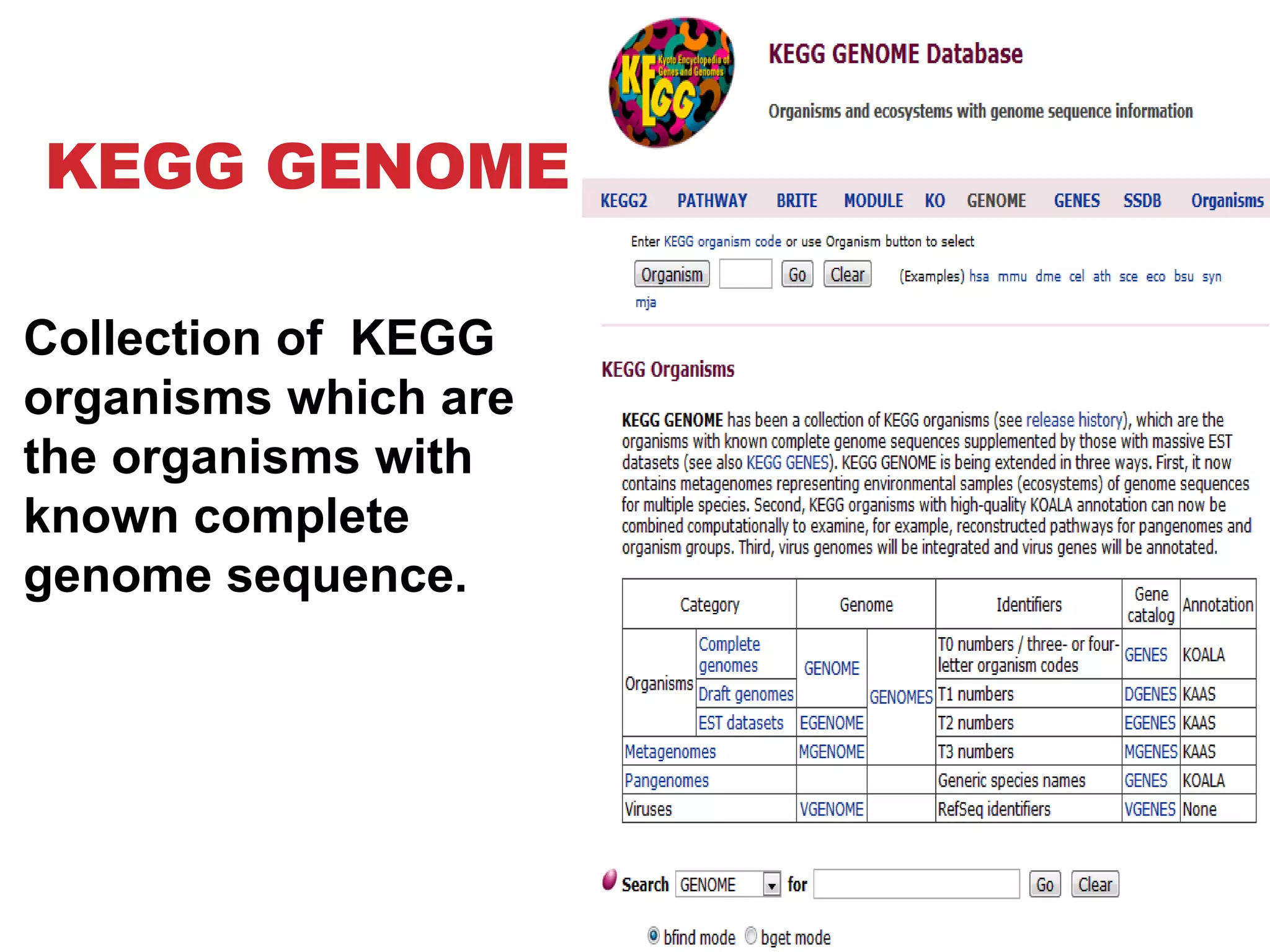
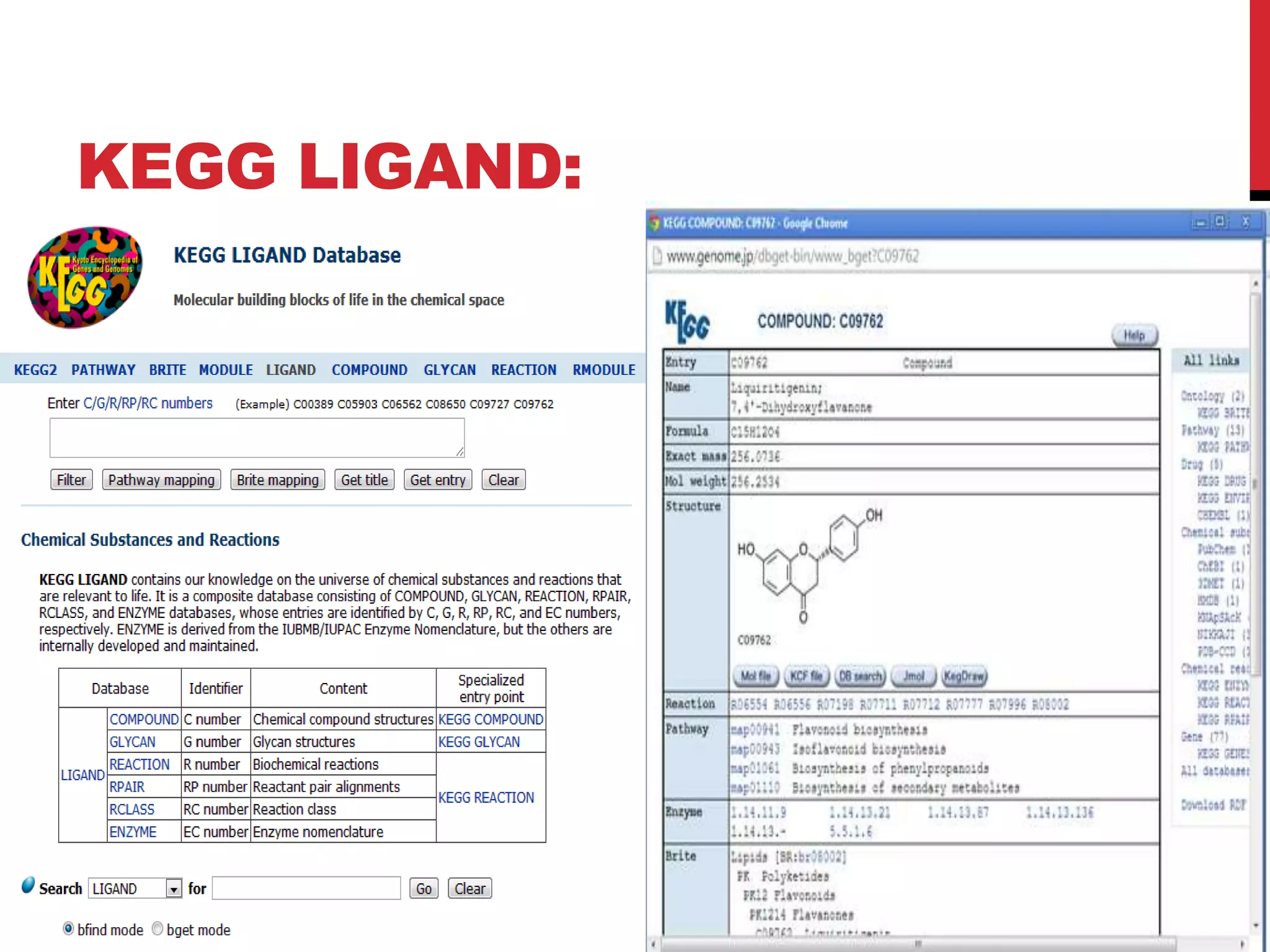
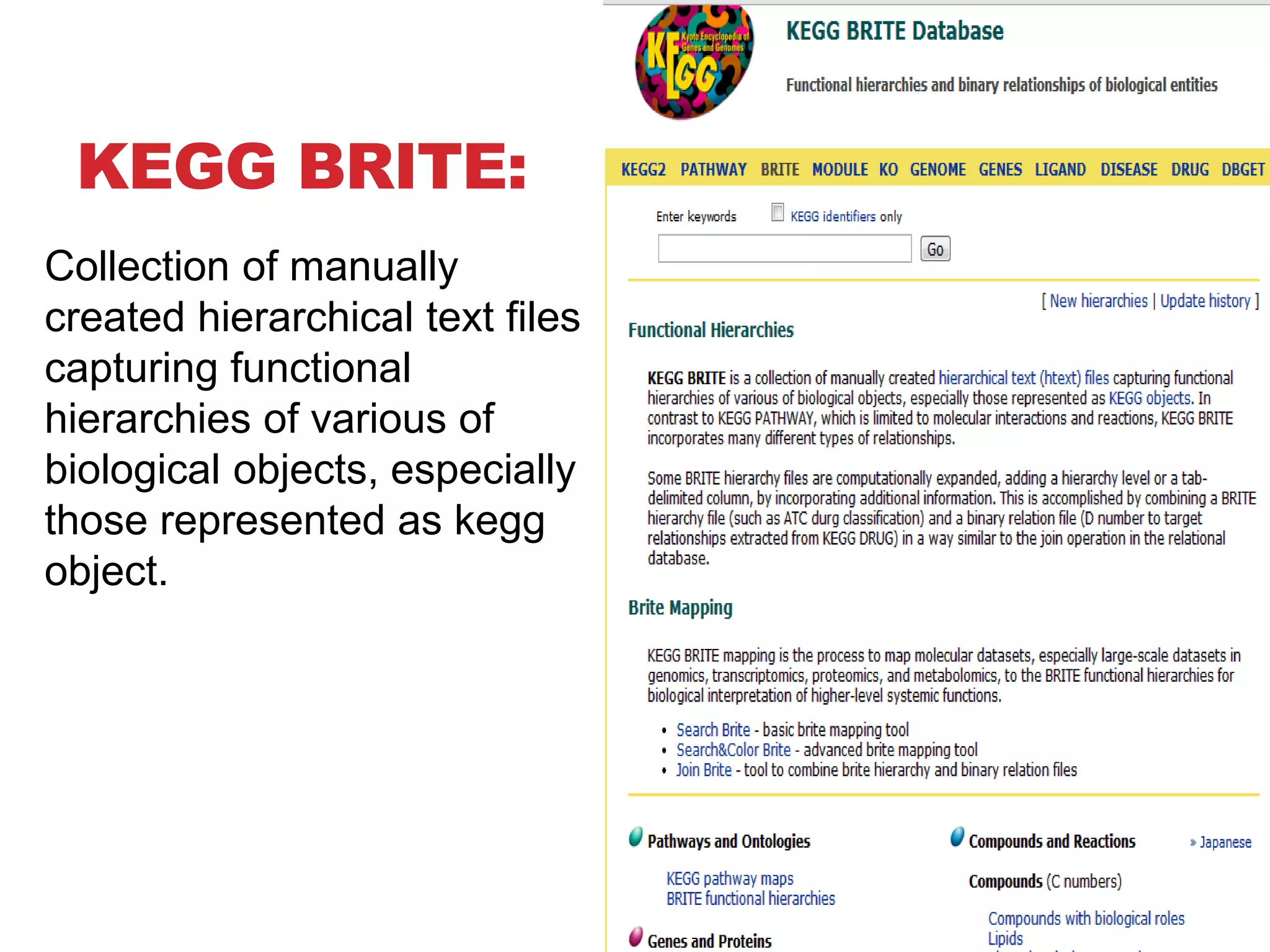
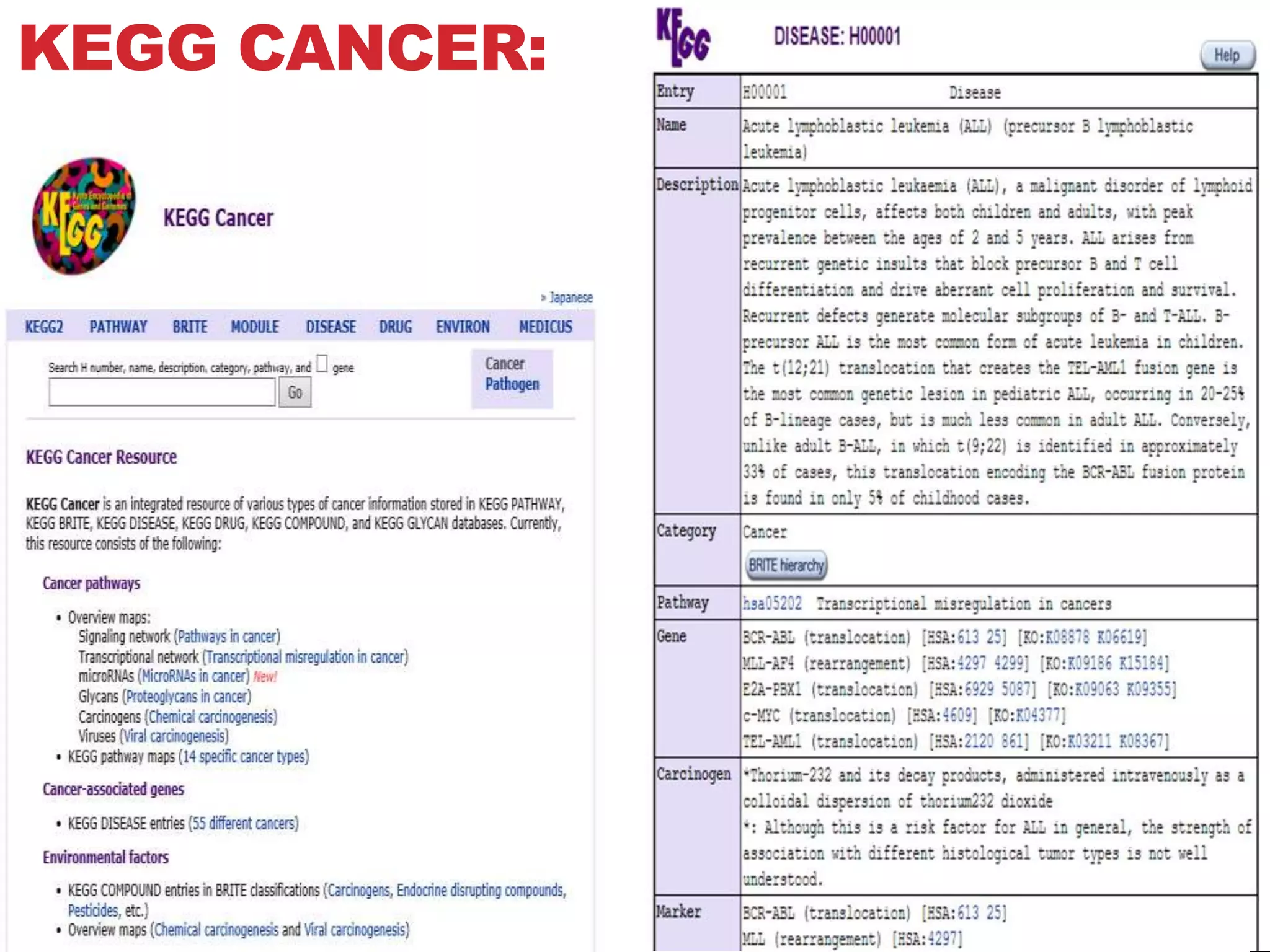
The document discusses the Kyoto Encyclopedia of Genes and Genomes (KEGG), which is an online database that contains information on genomes, enzymatic pathways, and biological chemicals. KEGG aims to computerize current biological knowledge and provide consistent annotations. It maintains six main databases: KEGG Pathway, KEGG Genes, KEGG Genome, KEGG Ligand, KEGG BRITE, and KEGG Cancer. These databases contain information on metabolic pathways, gene catalogs, genome sequences, chemical reactions, functional hierarchies, and cancer pathways respectively. KEGG can be used to detect enzyme clusters, compare gene clusters across genomes, and model and simulate biological systems.