Biological databases provide structured information on genes, proteins, and molecular structures. Major databases include:
- Sequence databases like NCBI and UniProt that contain nucleotide and protein sequences with associated metadata.
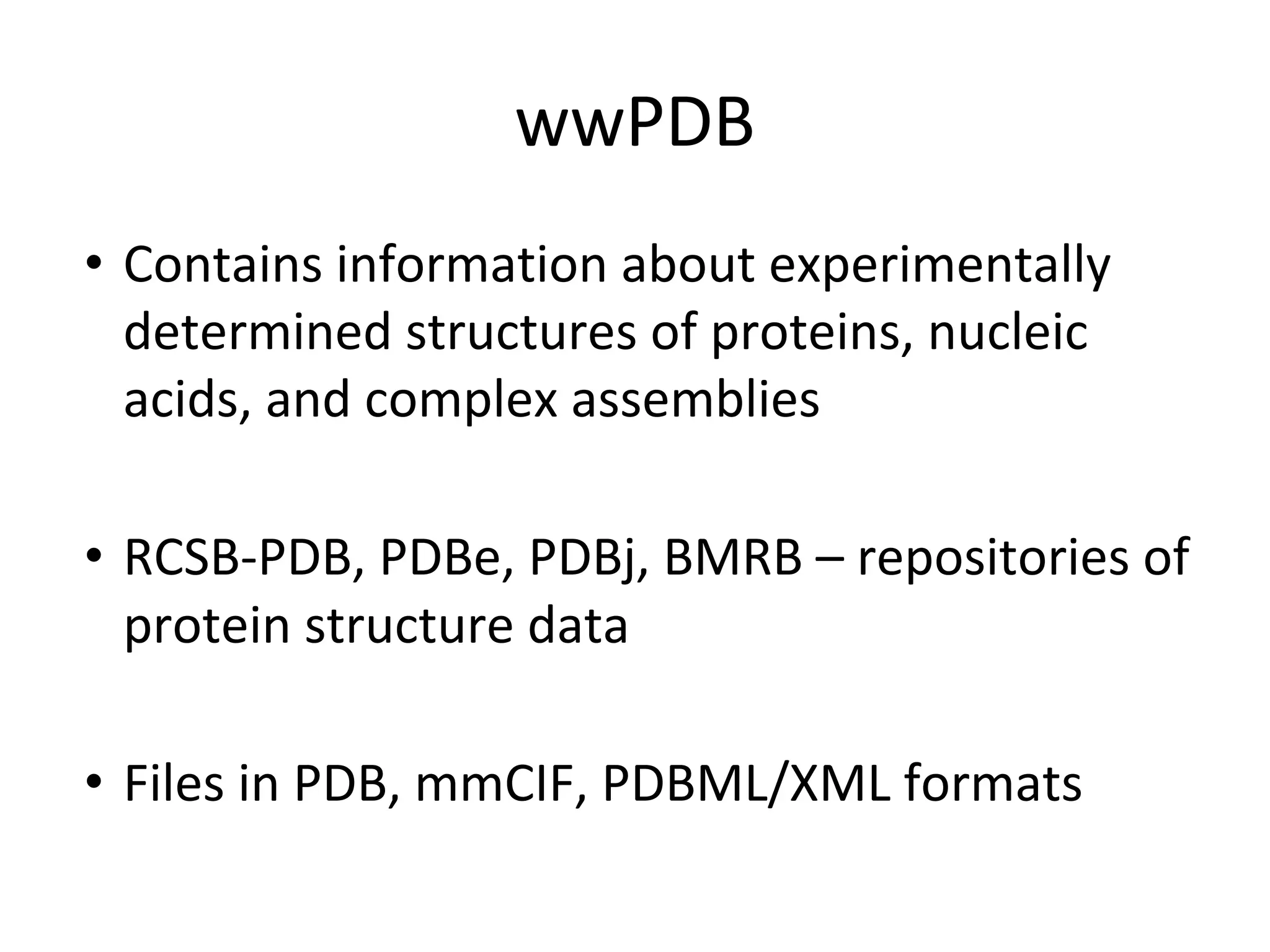
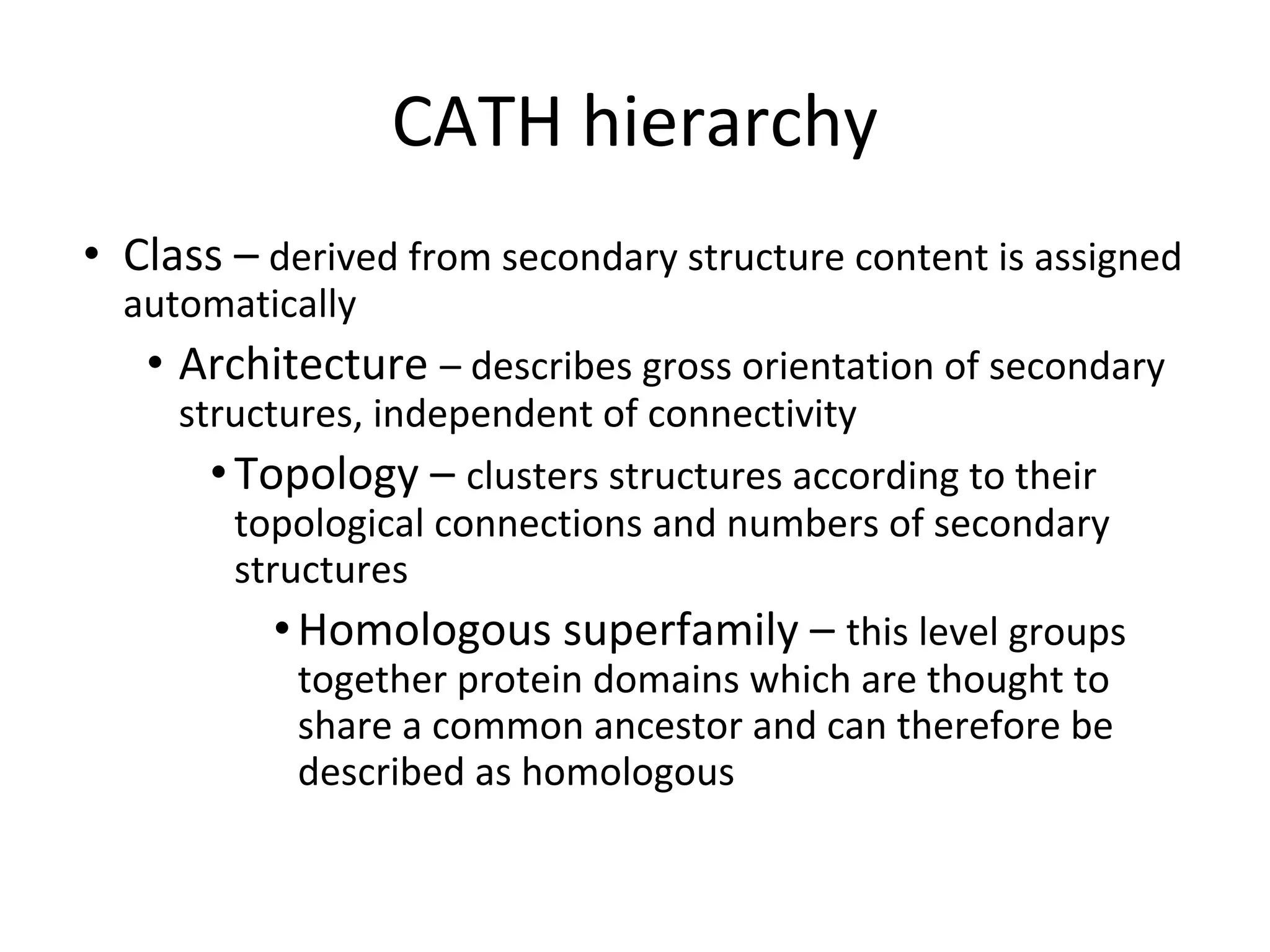
- Structure databases like PDB, CATH and SCOP that house 3D protein structure data and classify structures in hierarchical systems.
- Tools for analyzing sequences and structures, performing searches, and visualizing data are available on these sites. Biological databases aim to organize vast molecular information and link related data across resources.