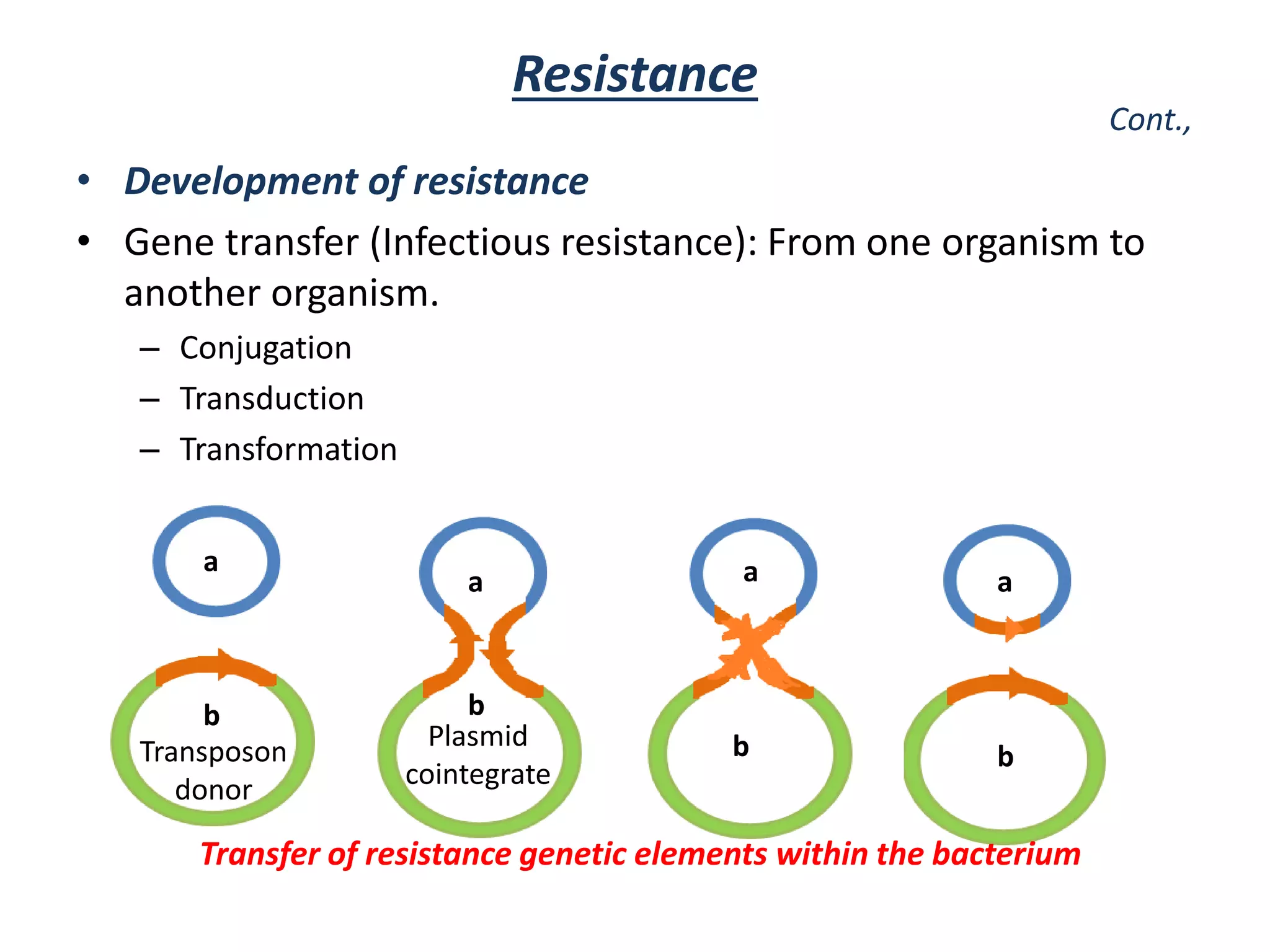
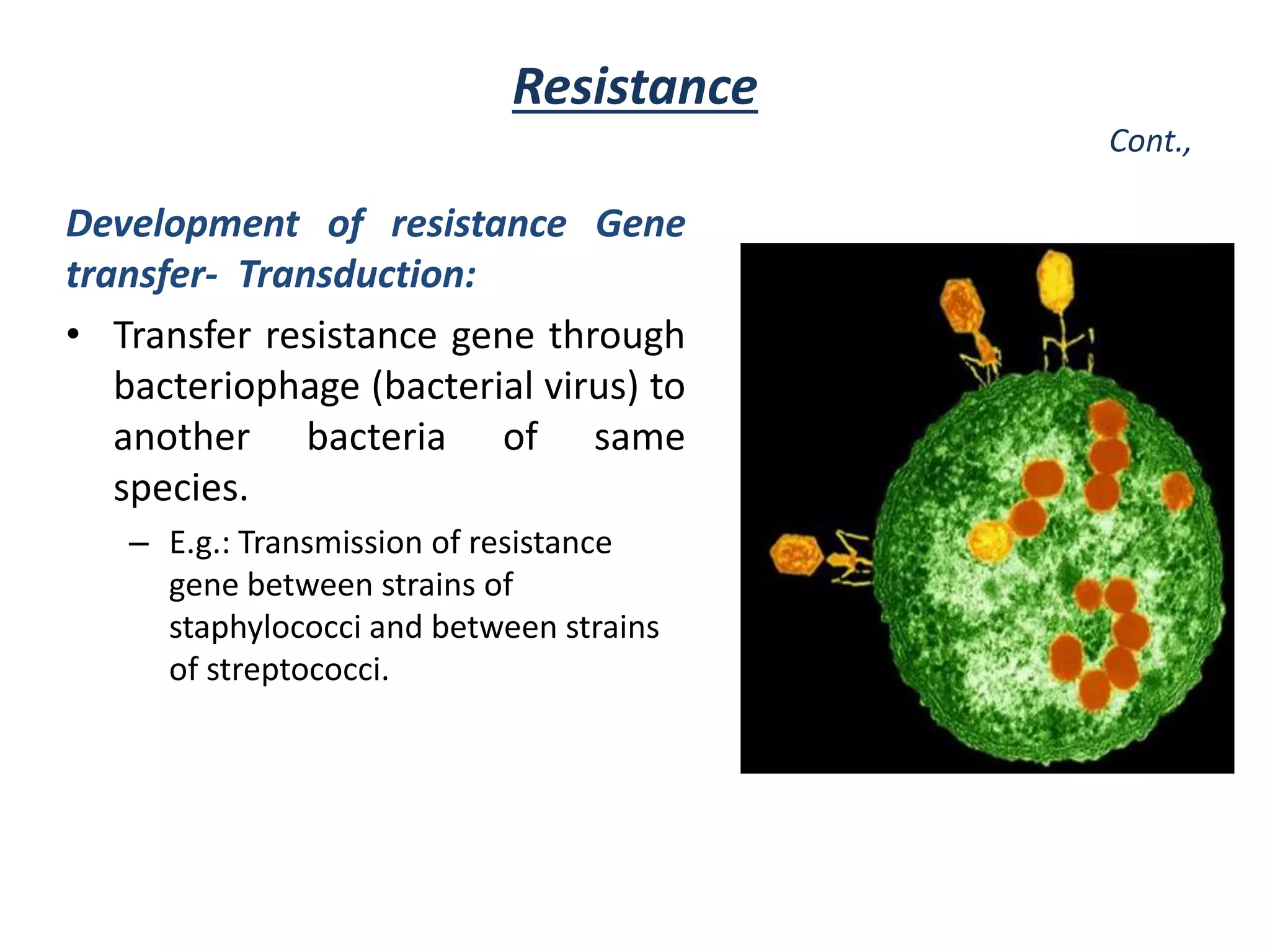
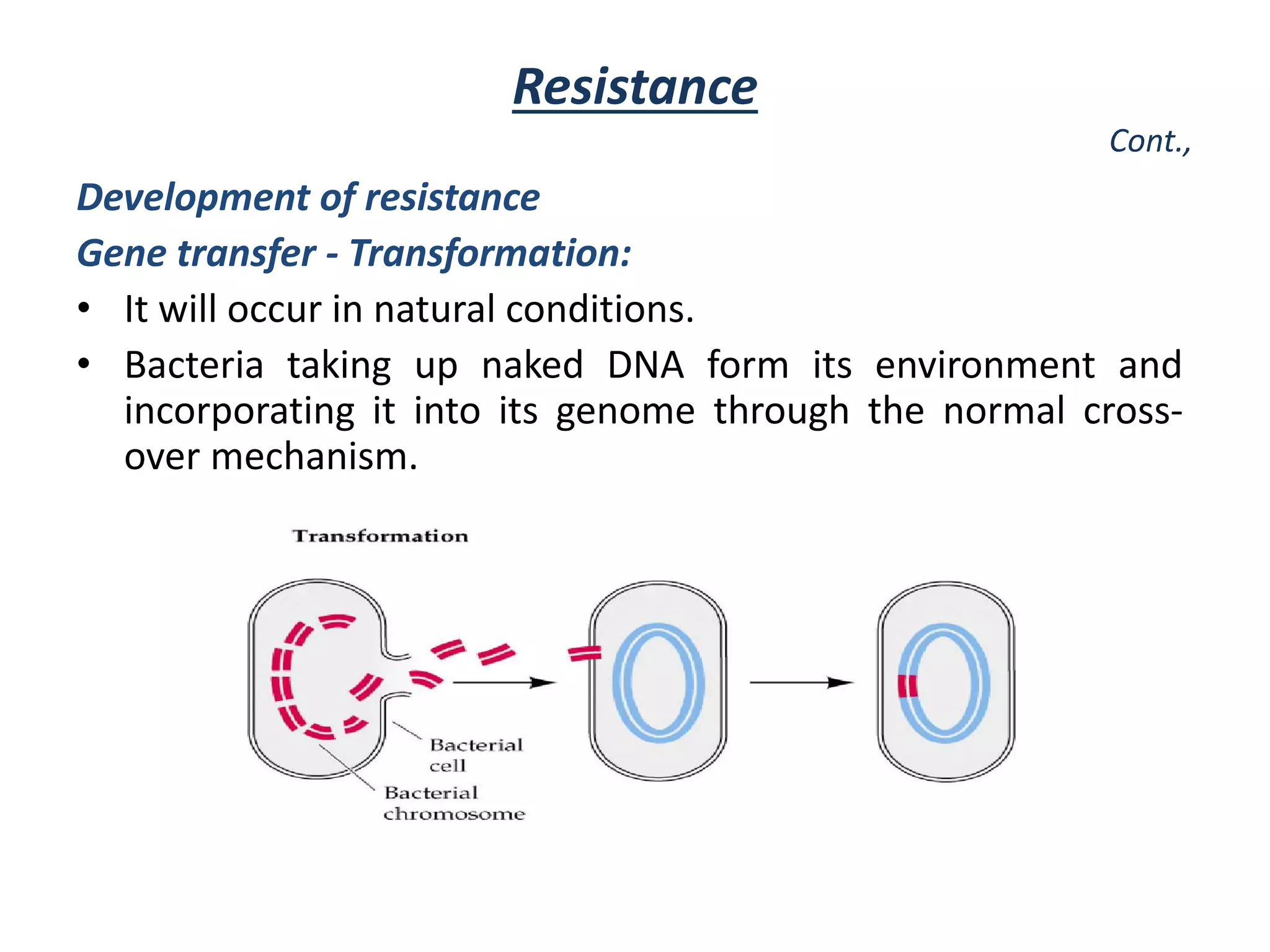
This document discusses the development of bacterial resistance to antimicrobials. It describes natural resistance, where some microbes are inherently resistant to certain antimicrobial medications (AMAs). It also describes acquired resistance, where microorganisms develop resistance over time due to widespread use of AMAs. Resistance can develop through mutation of genes or transfer of resistance genes between bacteria, such as through conjugation, transduction, or transformation. The widespread and long term use of certain AMAs has led to resistance developing in important pathogens like Staphylococcus, Mycobacterium tuberculosis, and Neisseria gonorrhoeae.