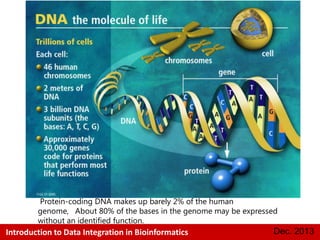
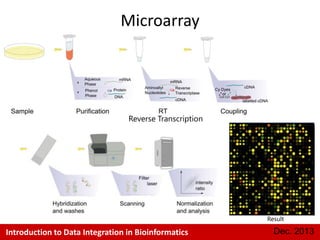
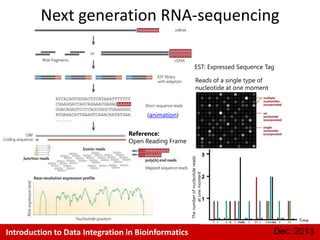
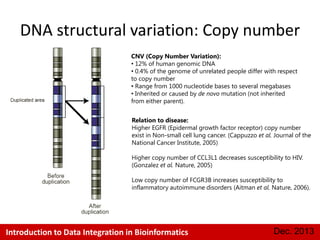
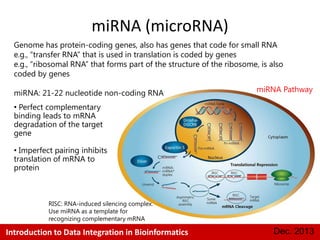
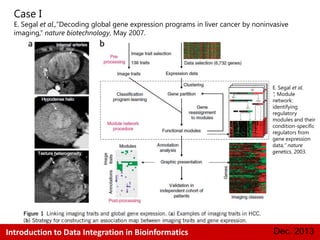
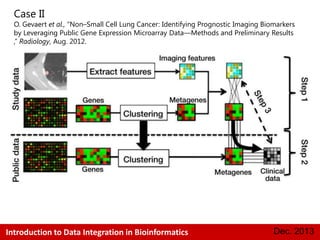
This document provides an introduction to data integration in bioinformatics. It discusses different types of biological data including gene expression, copy number variations, epigenetic data like DNA methylation, microRNA data, clinical data, and pathways. It also discusses challenges of data integration like data heterogeneity, incompleteness, and frequent changes. Finally, it provides examples of two case studies that integrate different types of biological and imaging data to study liver cancer and lung cancer.