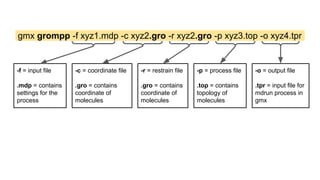
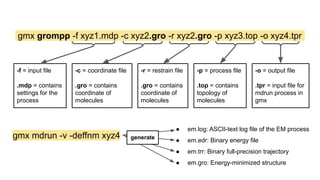
The document describes the steps to perform a molecular dynamics simulation of lysozyme in water using GROMACS. It details installing GROMACS in Google Colab, uploading input files, running energy minimization, equilibration, and production simulations. It also describes analyzing the trajectory by calculating the root mean square deviation (RMSD) and radius of gyration (Rg) to monitor stability of the protein structure over time.