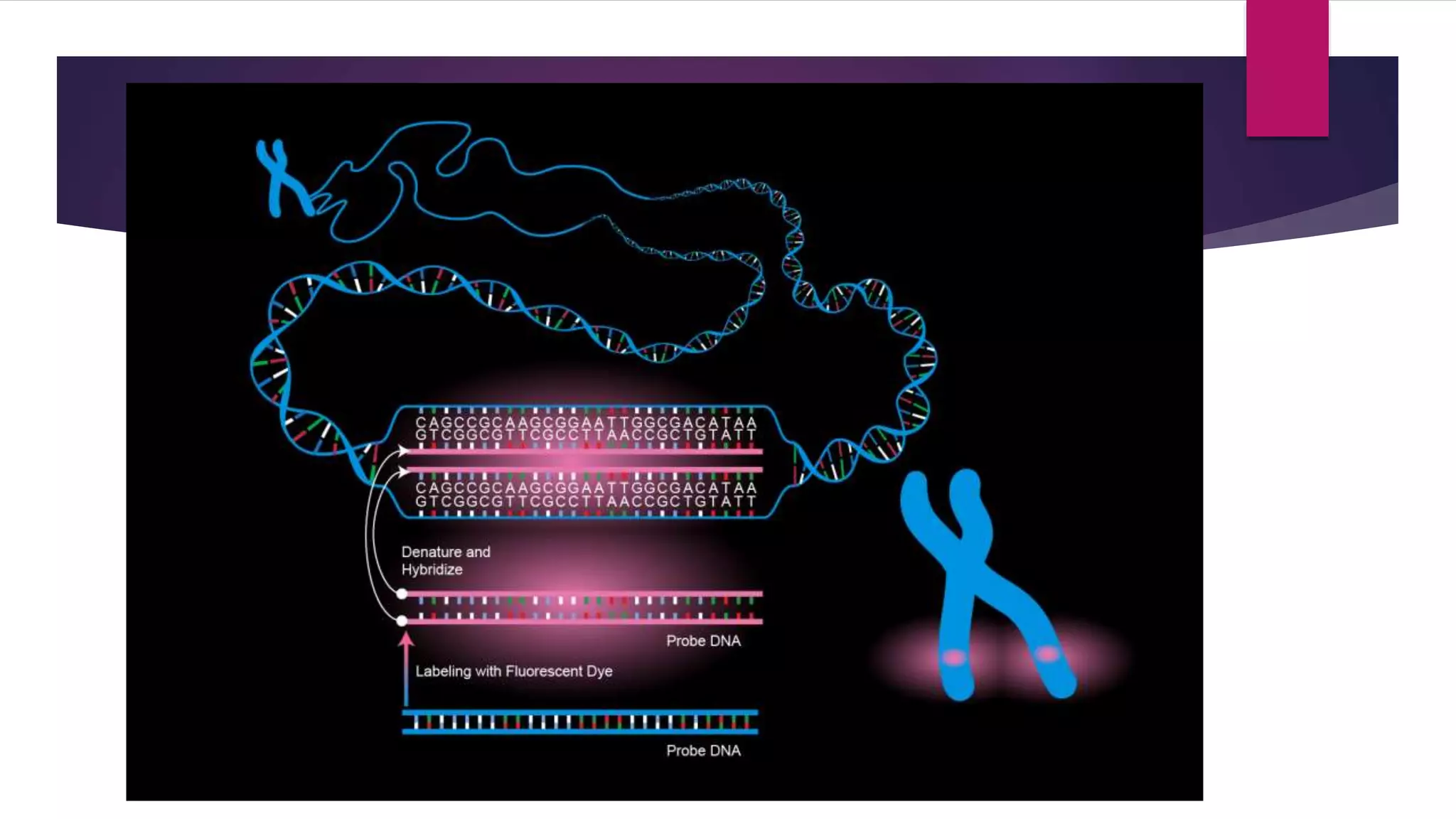
Genomic in-situ hybridization (GISH) is a cytogenetic technique developed for detecting and localizing specific nucleic acid sequences on chromosomes, using genomic DNA as probes. First established for animal cell hybrids in 1986, GISH was later adapted for plants by M.D. Bennett and involves several steps, including DNA extraction, slide preparation, and visualization under a fluorescence microscope. This method is valuable for identifying chromosomal rearrangements in cancer, studying genome evolution, and understanding the relationships of wild and cultivated plant species.