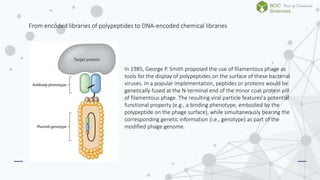
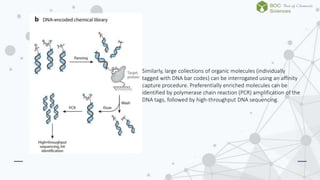
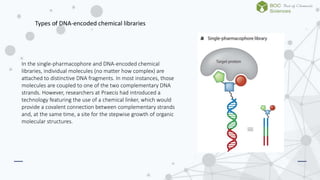
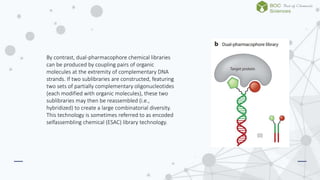
The document discusses DNA-encoded library technology, which enables the discovery of ligands through the use of filamentous phage for displaying polypeptides coupled with genetic information. It outlines the historical development of this technology from initial proposals in the 1980s to current applications in generating large libraries of chemical compounds with DNA barcoding for affinity selection. Additionally, it describes variations in library types, including single- and dual-pharmacophore libraries that enhance combinatorial diversity.