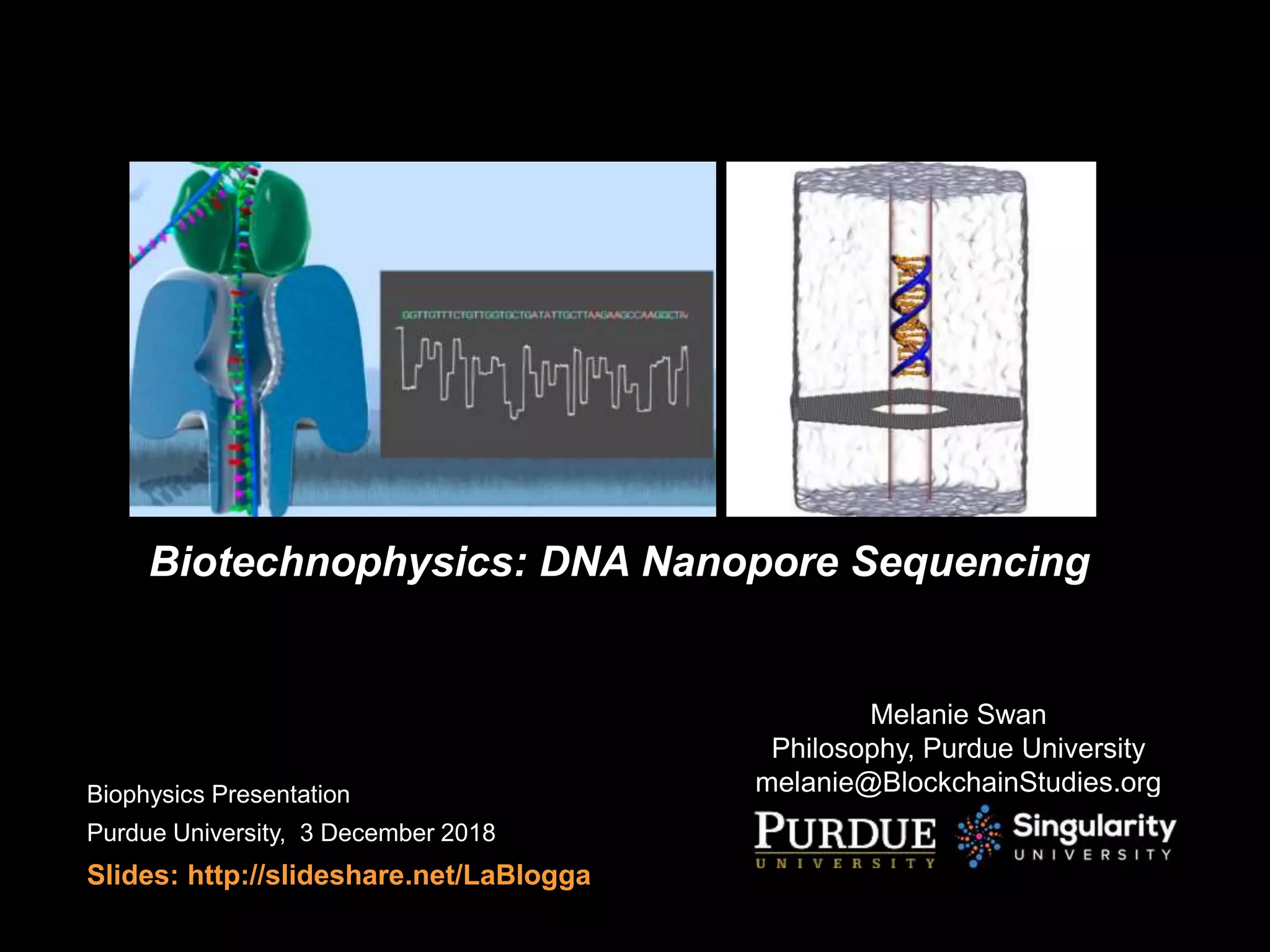
The document presents a detailed exploration of DNA nanopore sequencing and its significance in biotechnophysics, emphasizing the need for a biophysical understanding to advance biotechnological applications. It outlines the evolution of DNA sequencing platforms, from Sanger to contemporary nanopore technologies, and discusses the mechanisms affecting DNA capture rates in nanopores. Various simulation results highlight the complex interplay of electric fields and water dynamics that can influence biomolecule transport through nanopore systems.