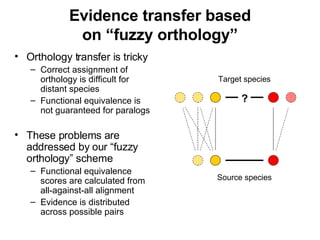
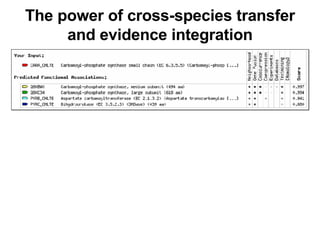
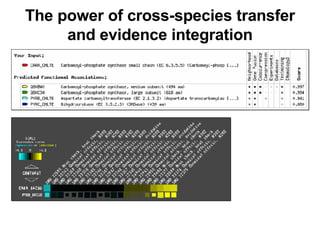
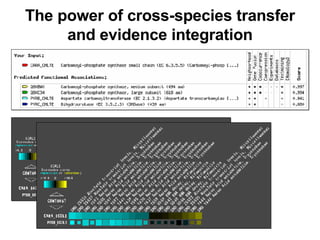
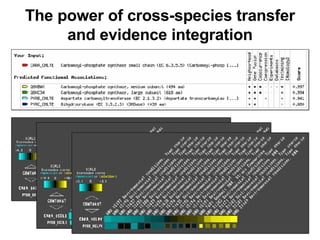
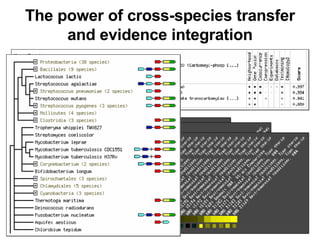
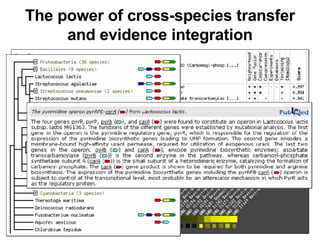
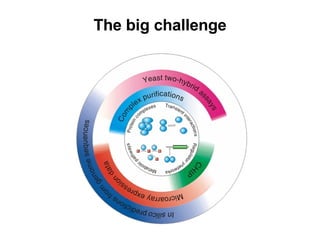
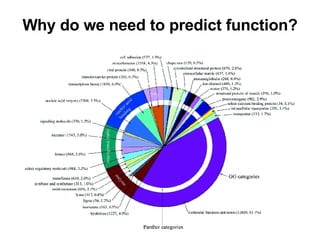
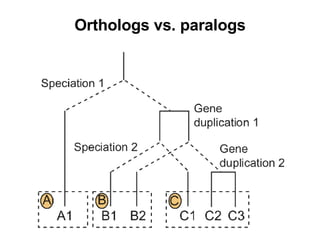
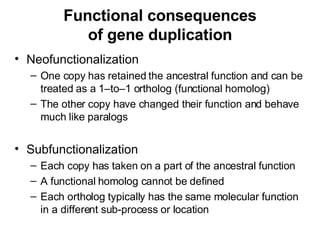
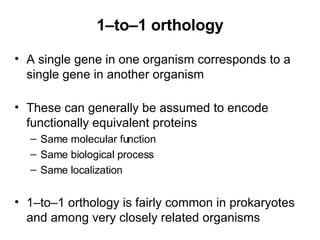
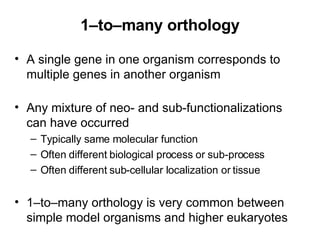
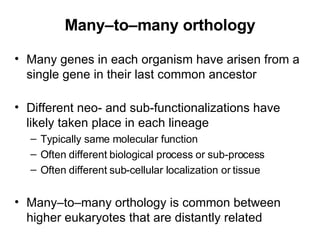
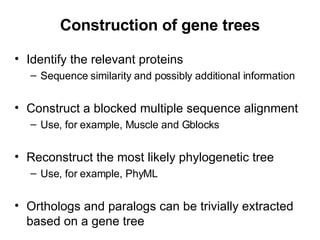
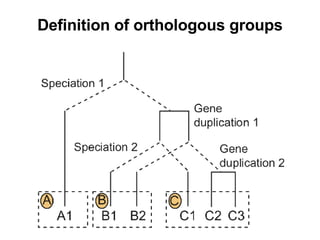
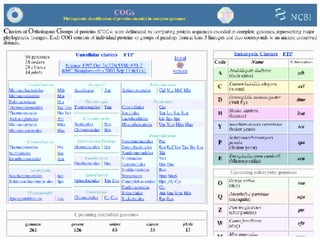
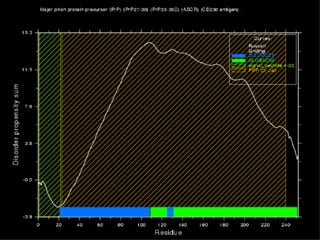
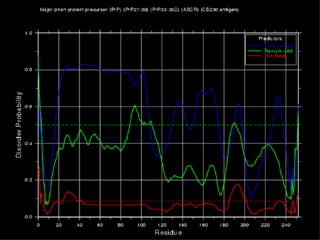
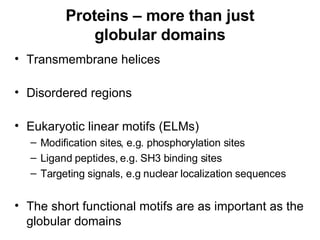
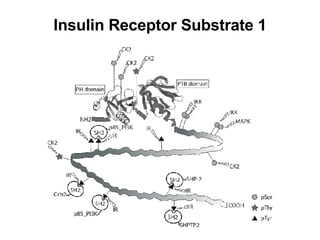
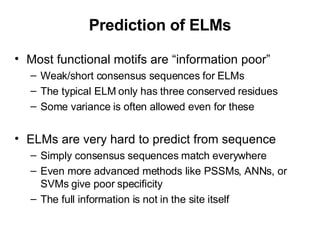
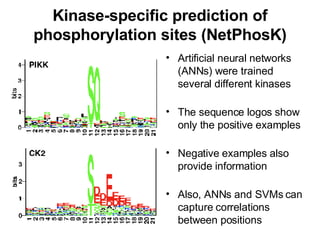
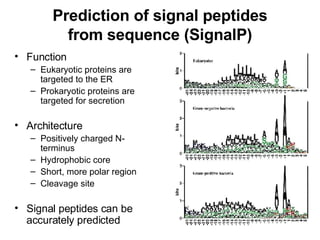
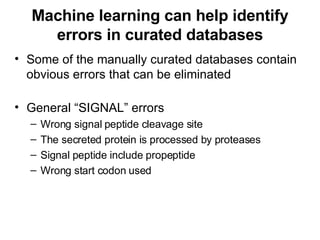
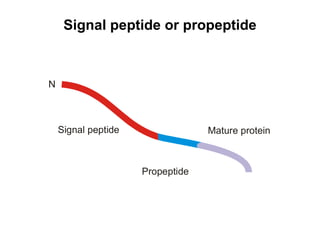
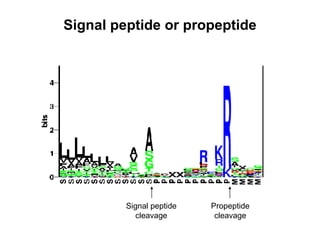
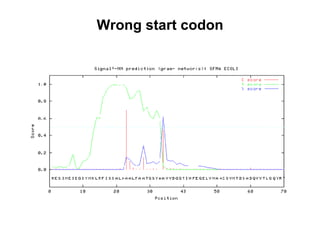
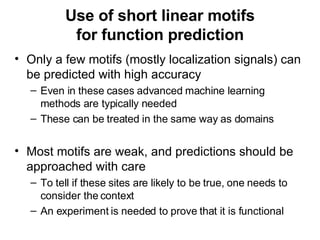
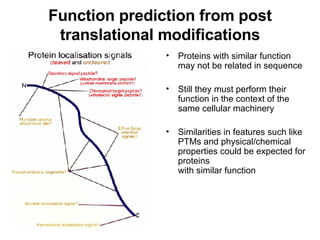
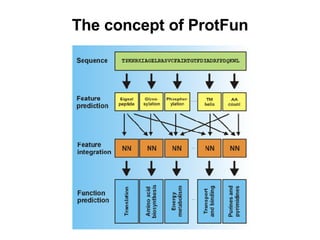
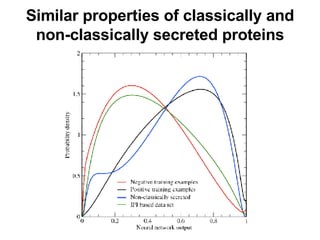
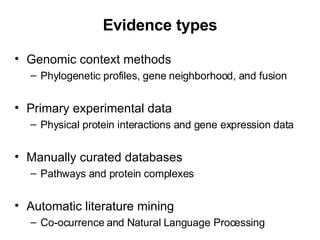
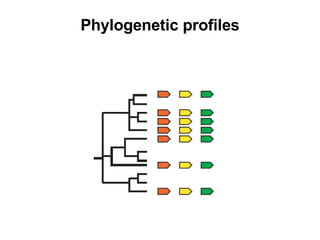
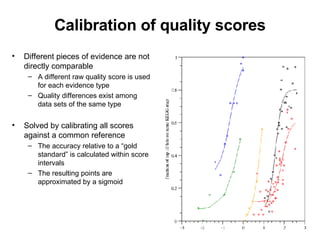
The document discusses various methods for predicting protein function, including homology-based transfer of annotation and prediction of functional motifs and domains. Homology-based transfer can infer molecular function from sequence similarity, but biological process is only transferable between orthologs. Orthologs can be detected through phylogenetic trees or automated methods like InParanoid. Each protein domain contributes to molecular function, while short motifs like phosphorylation sites are also important. Functional annotation involves describing proteins at the molecular, biological process, and cellular component levels.






















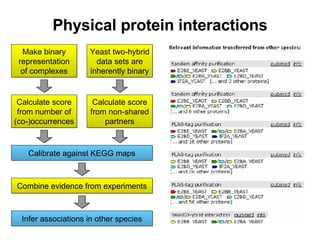
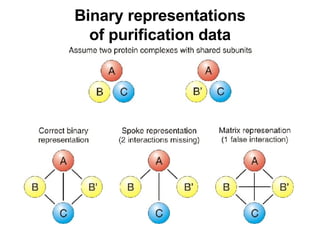
![Topology based quality scores Scoring scheme for yeast two-hybrid data: S1 = -log((N 1 +1) · (N 2 +1)) N 1 and N 2 are the numbers of non-shared interaction partners Similar scoring schemes have been published by Saito et al. Scoring scheme for complex pull-down data: S2 = log[(N 12 · N)/((N 1 +1) · (N 2 +1))] N 12 is the number of purifications containing both proteins N 1 is the number containing protein 1, N 2 is defined similarly N is the total number of purifications Both schemes aim at identifying ubiquitous interactors](https://image.slidesharecdn.com/jen06talk10-1217971785207872-9/85/Protein-function-prediction-126-320.jpg)
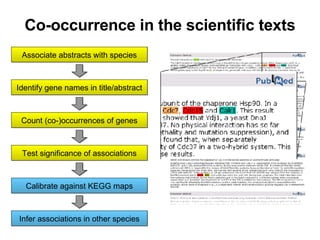
![Gene and protein names Cue words for entity recognition Verbs for relation extraction [ nxgene The GAL4 gene ] [ nxexpr T he expression of [ nxgene the cytochrome genes [ nxpg CYC1 and CYC7 ]]] is controlled by [ nxpg HAP1 ] Natural Language Processing](https://image.slidesharecdn.com/jen06talk10-1217971785207872-9/85/Protein-function-prediction-131-320.jpg)