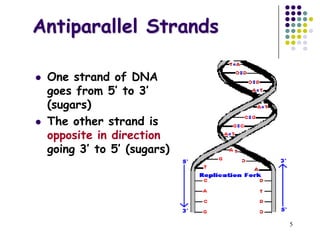
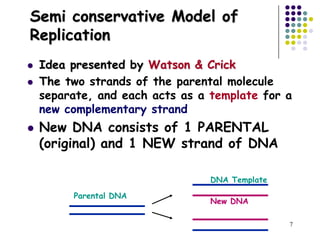
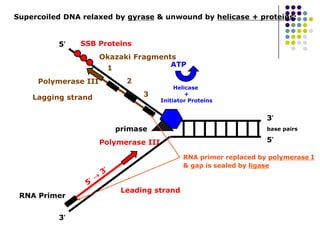
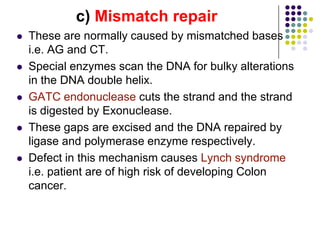
The document discusses DNA replication and repair, emphasizing the flow of genetic information and the structure of DNA. It explains the semi-conservative model of DNA replication, detailing the roles of enzymes like DNA helicase, primase, and polymerases in synthesizing new DNA strands, as well as the necessity of RNA primers. Additionally, it covers various DNA repair mechanisms to correct replication errors and damage from environmental factors.