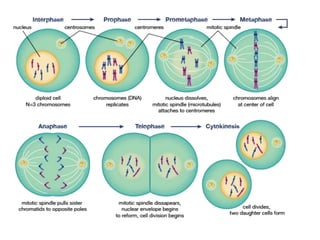
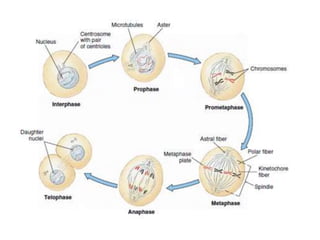
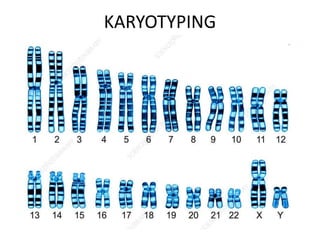
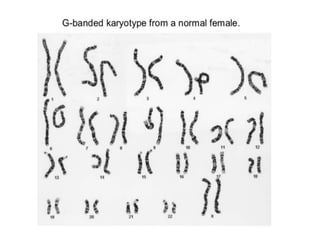
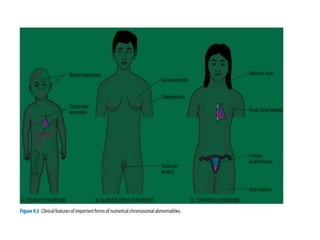
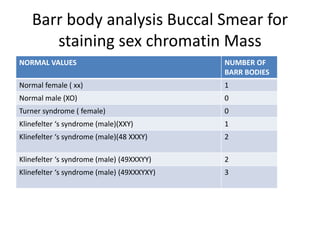
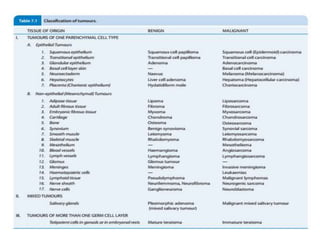
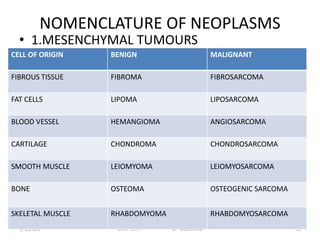
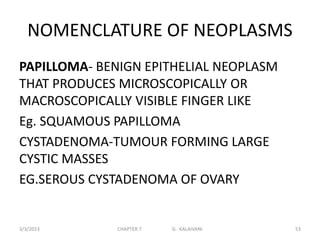
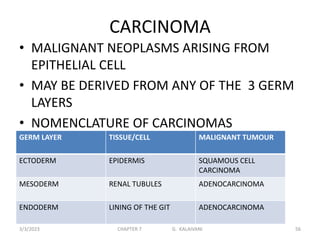
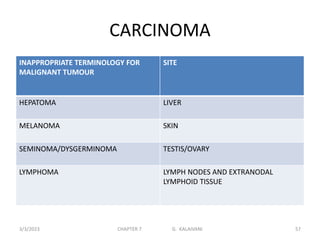
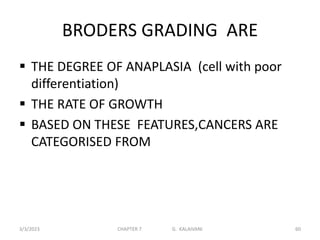
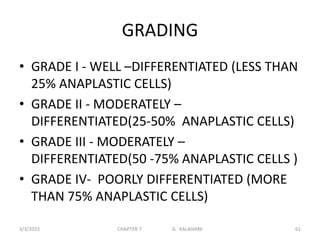
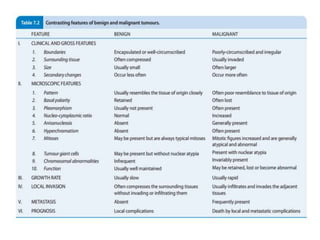
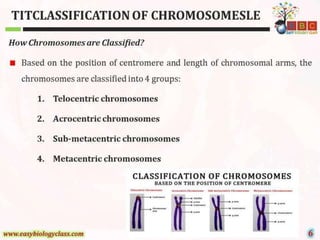
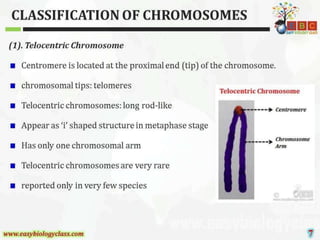
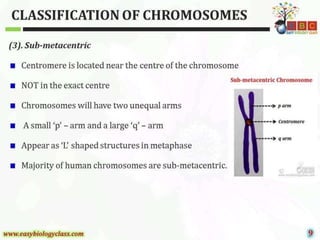
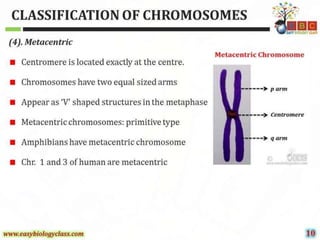
This document discusses cytogenetics and chromosome analysis techniques. It begins with an introduction to human chromosomes and chromosomal abnormalities. It then describes various types of chromosomal mutations and abnormalities that can be detected through karyotyping and fluorescence in situ hybridization (FISH). The document provides detailed procedures for chromosome sample preparation from bone marrow and blood cultures, as well as staining and analysis techniques like Giemsa staining and G-banding. The importance of chromosomal studies for diagnosing conditions like Turner syndrome and Klinefelter syndrome is also highlighted.