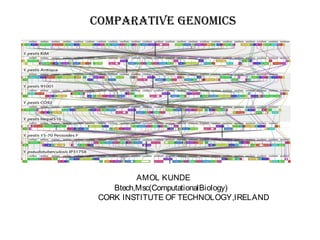
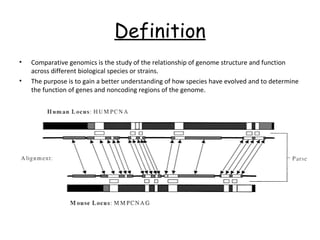
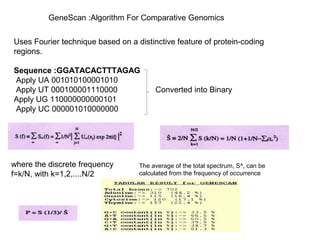
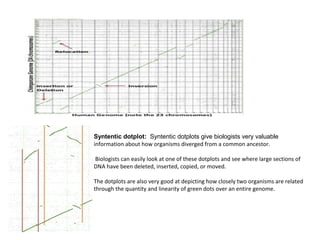
Comparative genomics is the study of genome structure and function across species. Sequencing entire genomes is non-optimal due to vast numbers of species and large genome sizes, and individuals within a species have genetically distinct genomes. Comparative genomics addresses these issues using approaches like gene prediction algorithms that use features of protein-coding regions and tools that find putative genes or syntenic regions between genomes.