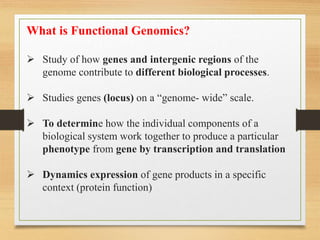
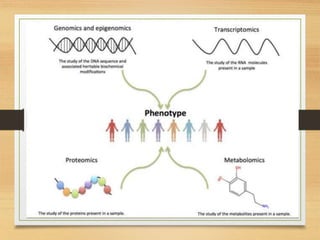
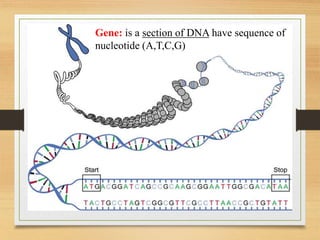
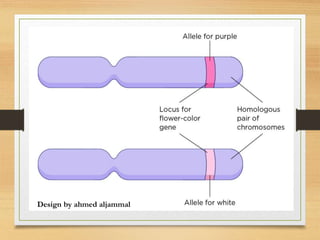
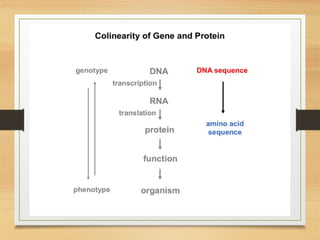
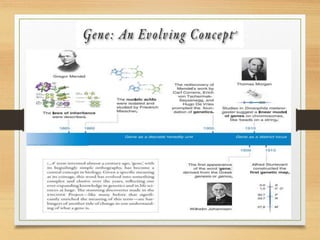
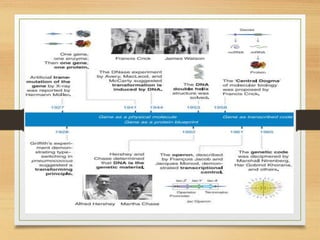
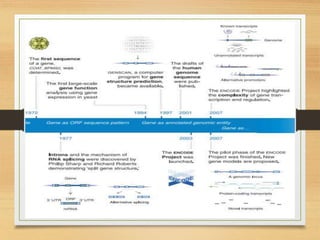
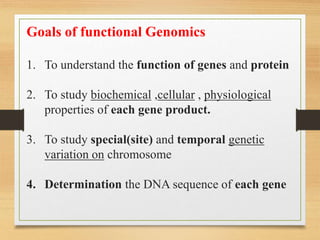
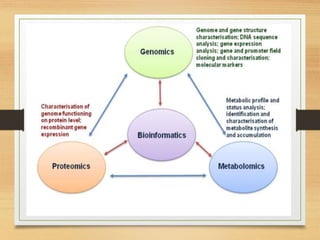
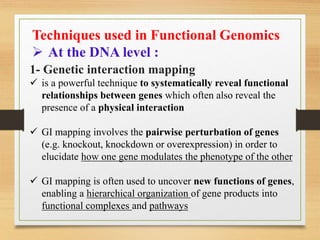
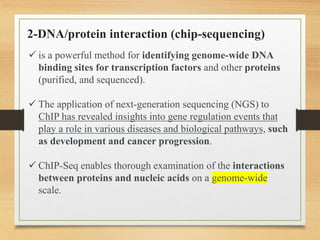
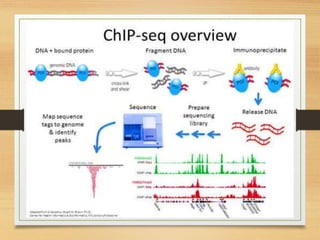
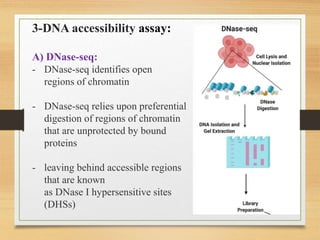
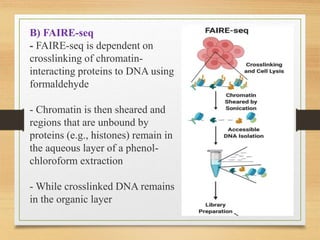
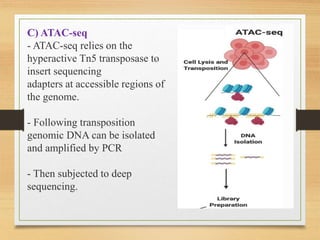
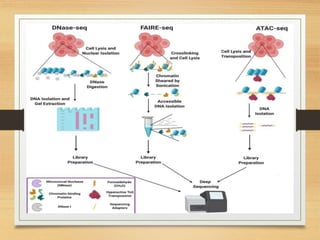
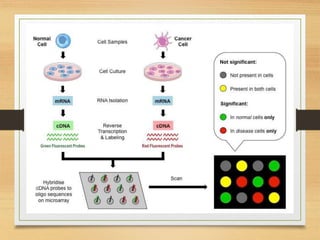
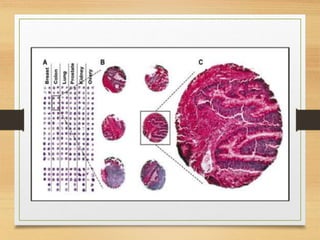
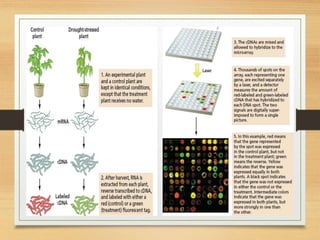
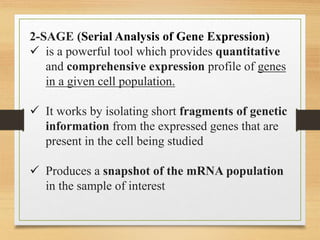
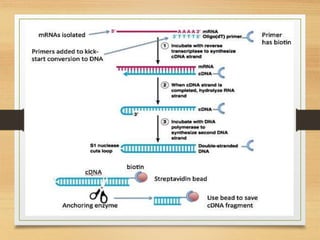
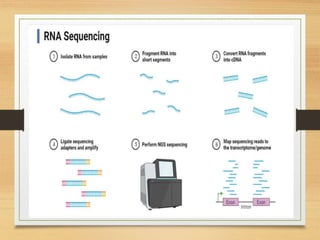
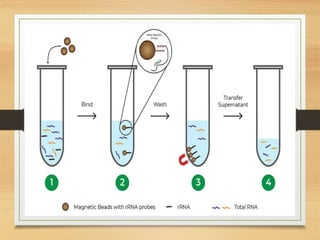
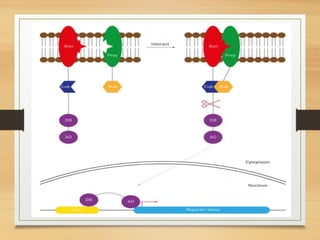
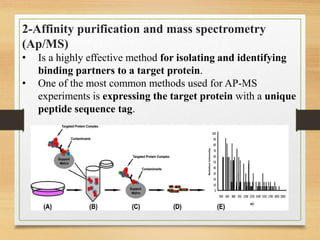
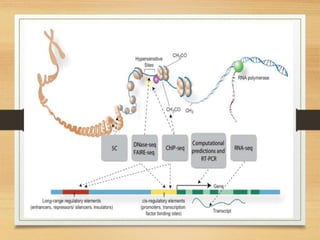
This document discusses functional genomics, which studies how genes and intergenic regions contribute to biological processes on a genome-wide scale. It describes techniques used at the DNA, RNA, and protein levels, including genetic interaction mapping, ChIP-seq, DNase-seq, FAIRE-seq, ATAC-seq, microarrays, SAGE, RNA-seq, yeast two-hybrid systems, and affinity purification mass spectrometry. Major projects like ENCODE and GTEx aim to identify all functional elements in genomic DNA and understand the role of genetic variation in gene expression across tissues.