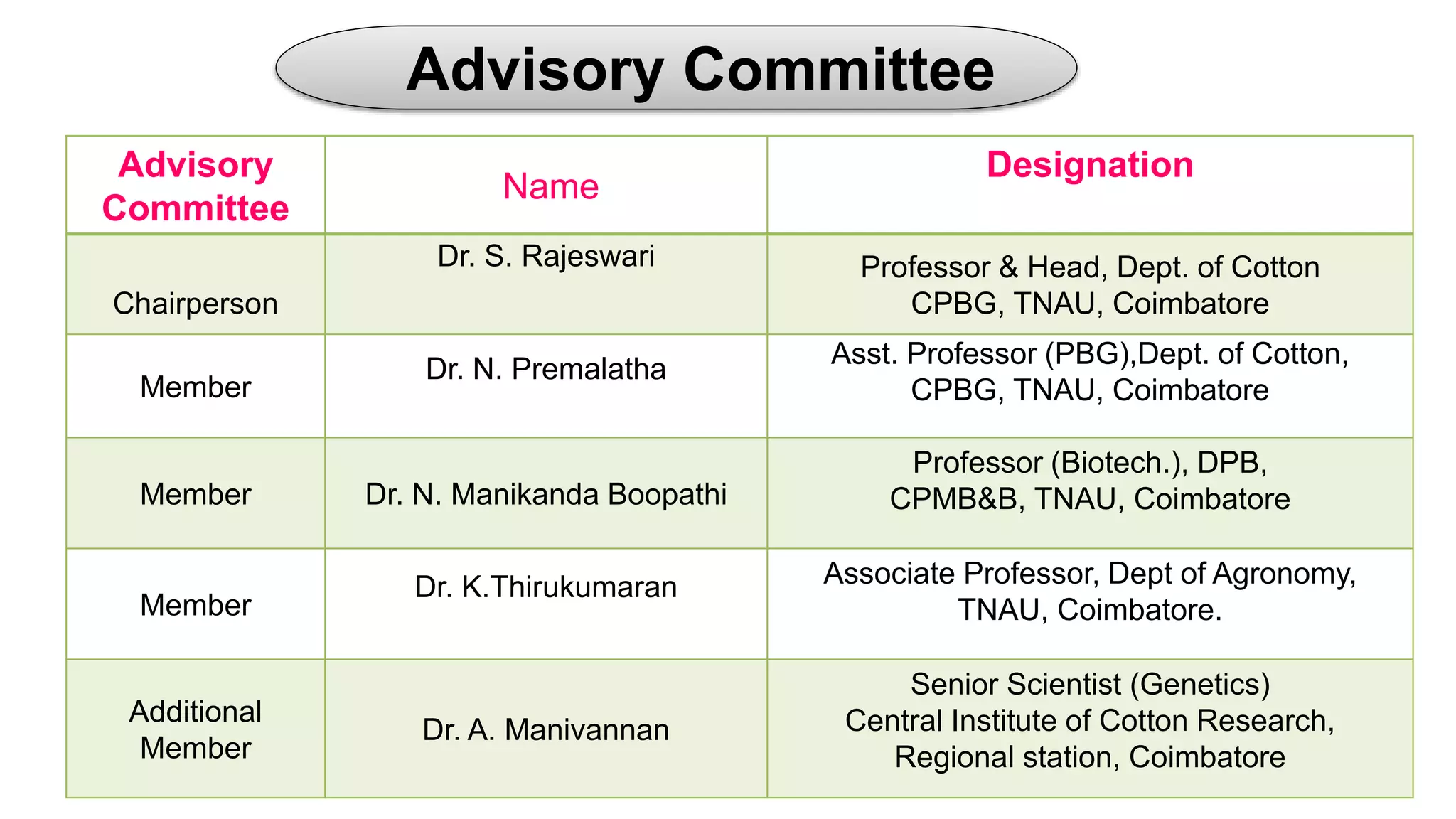
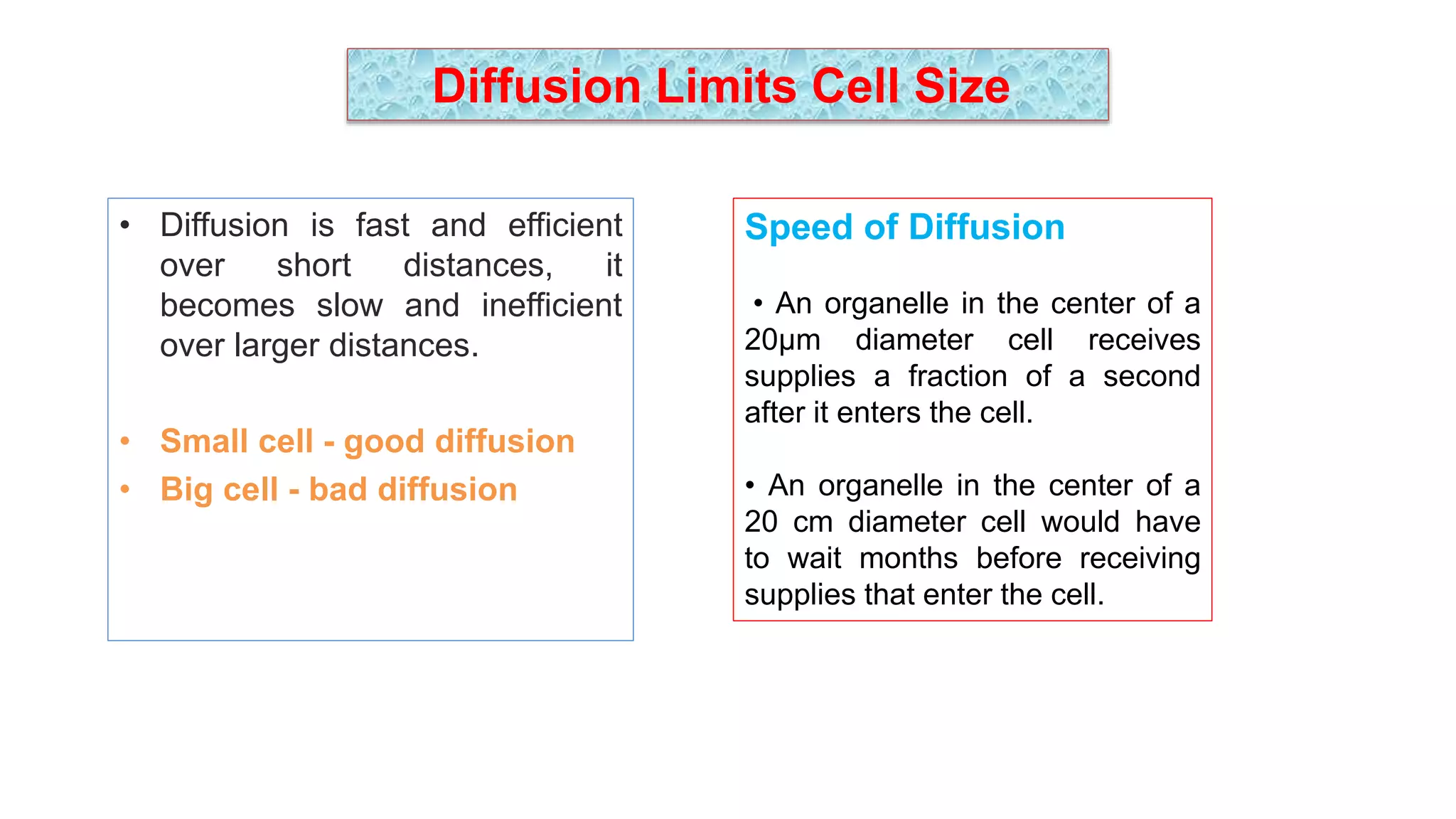
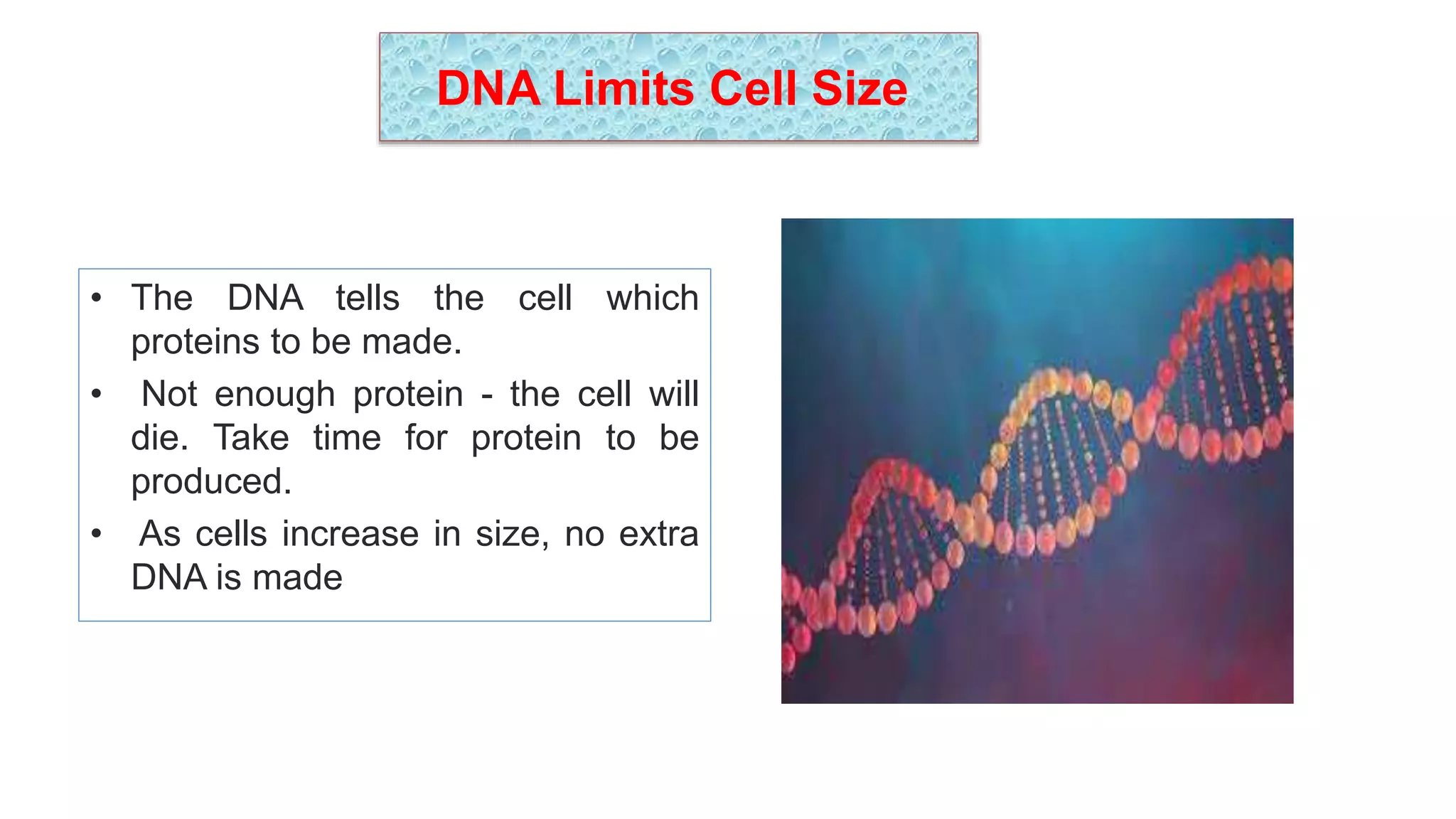
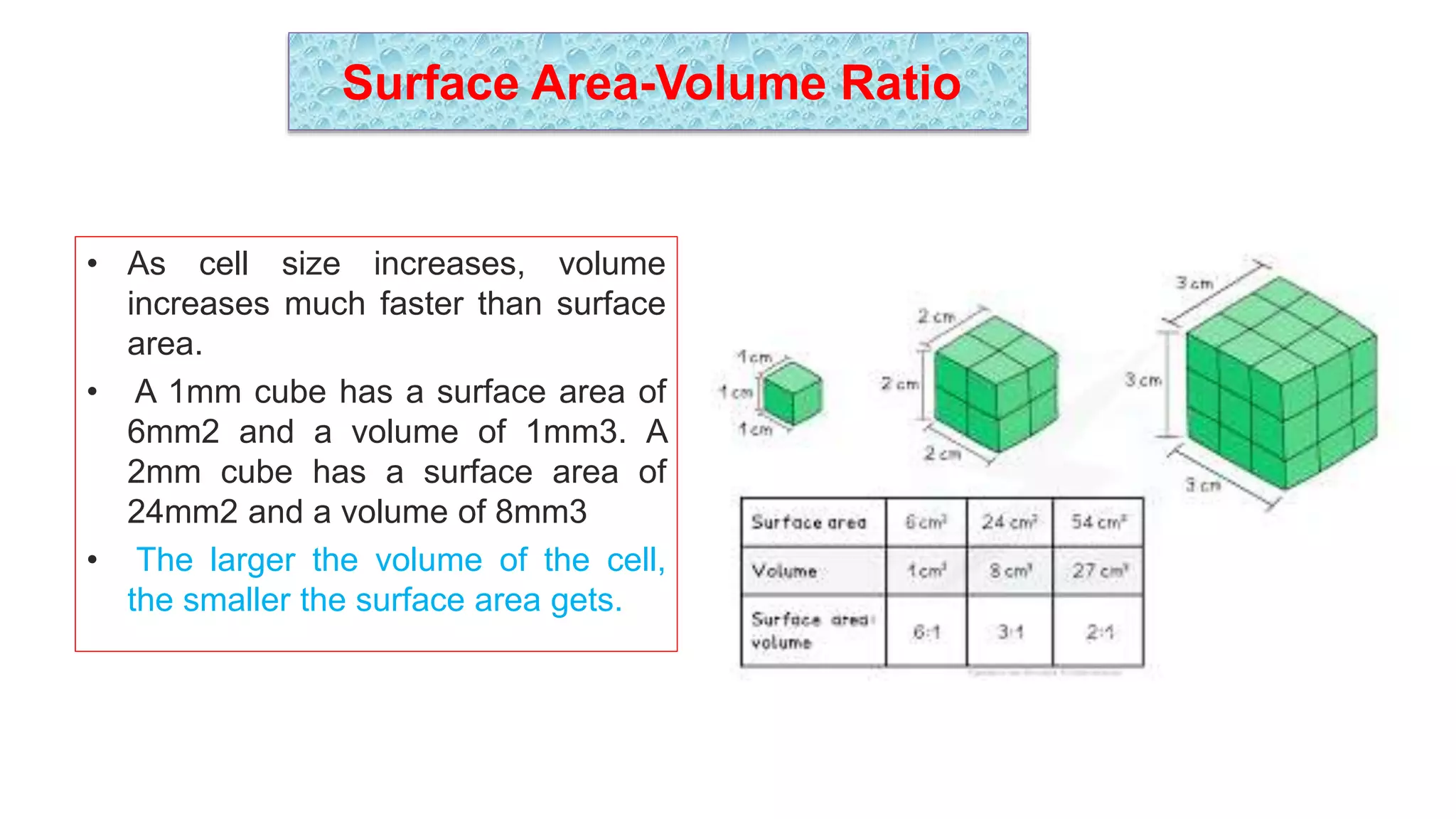
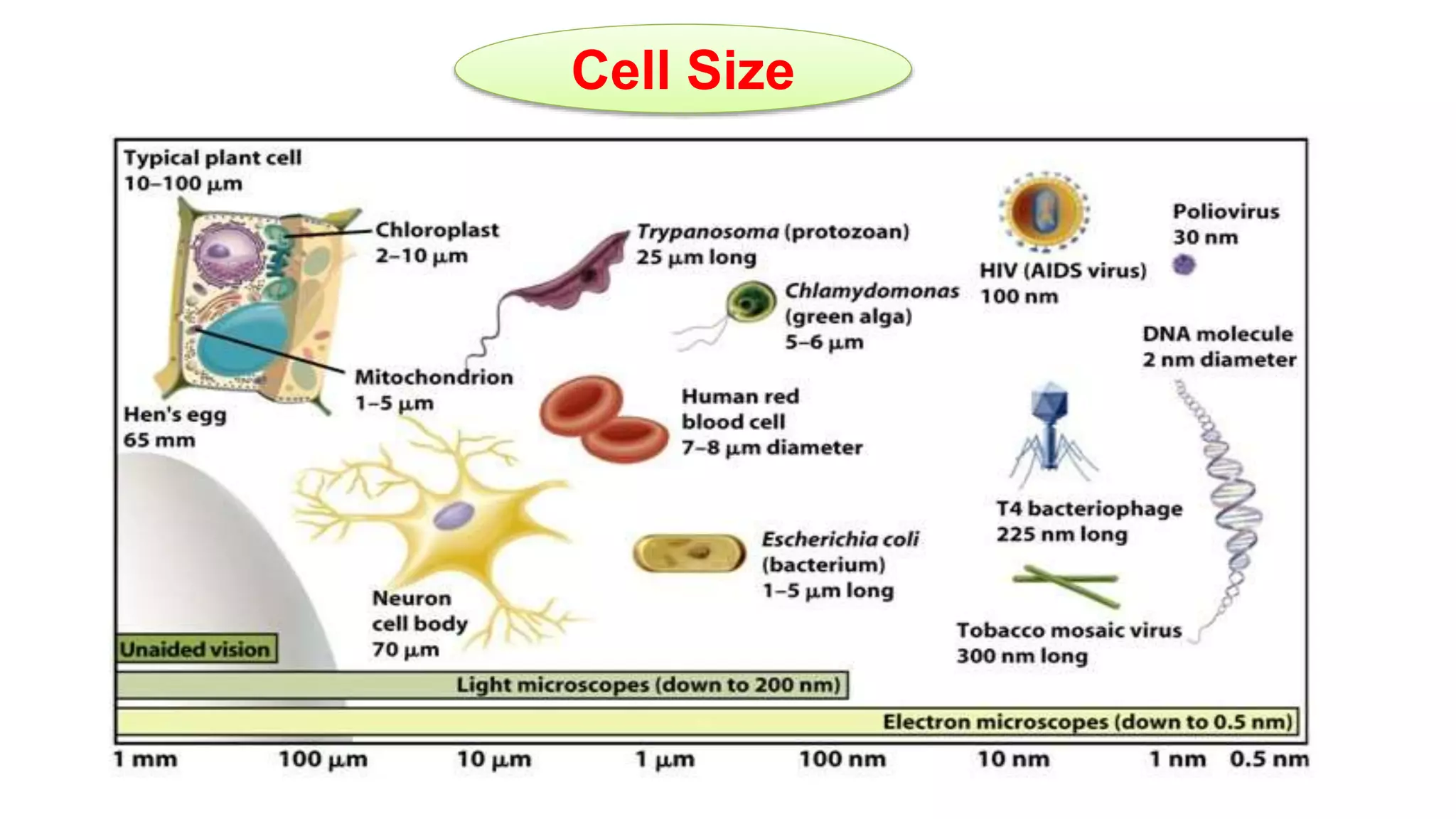
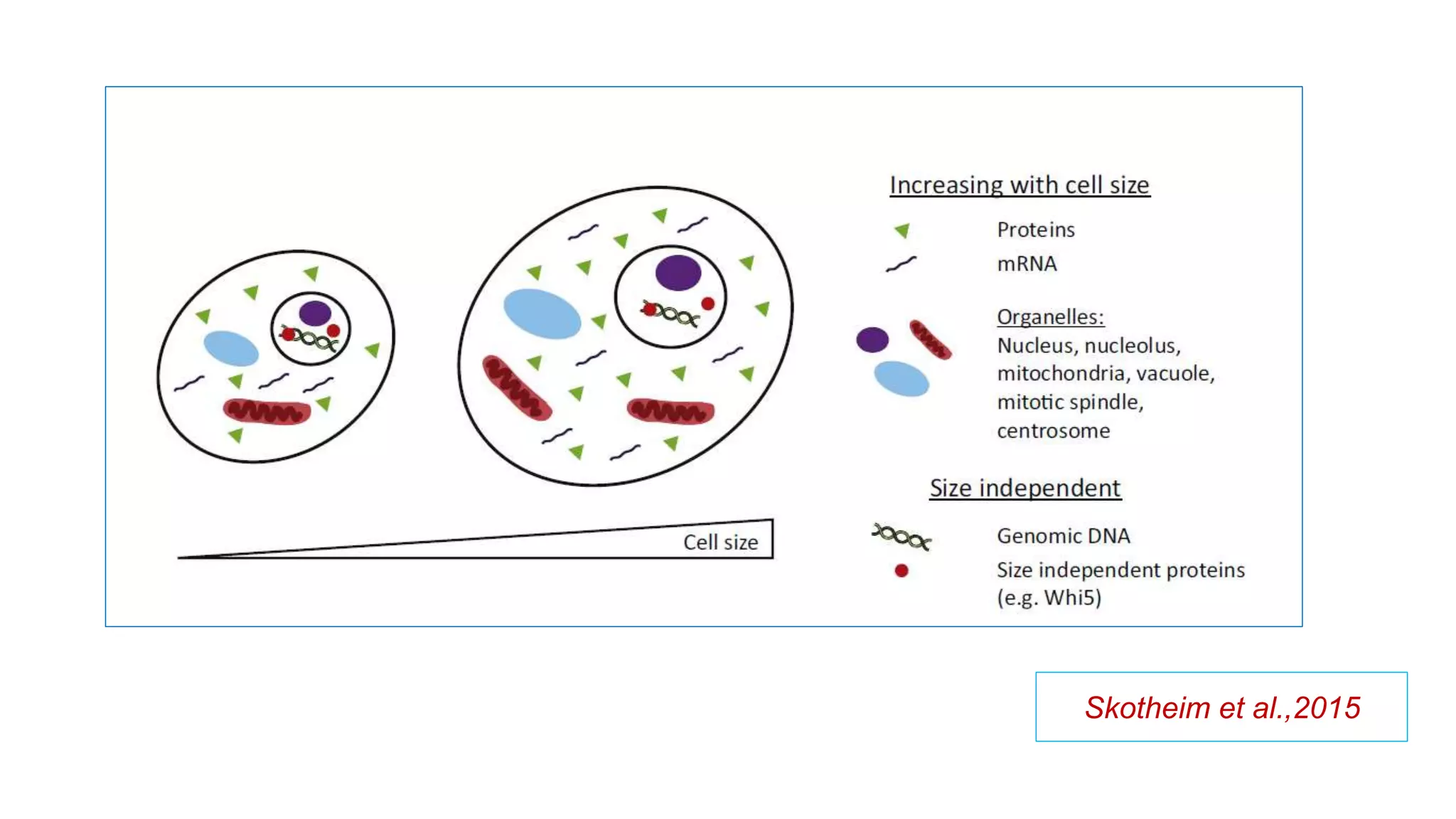
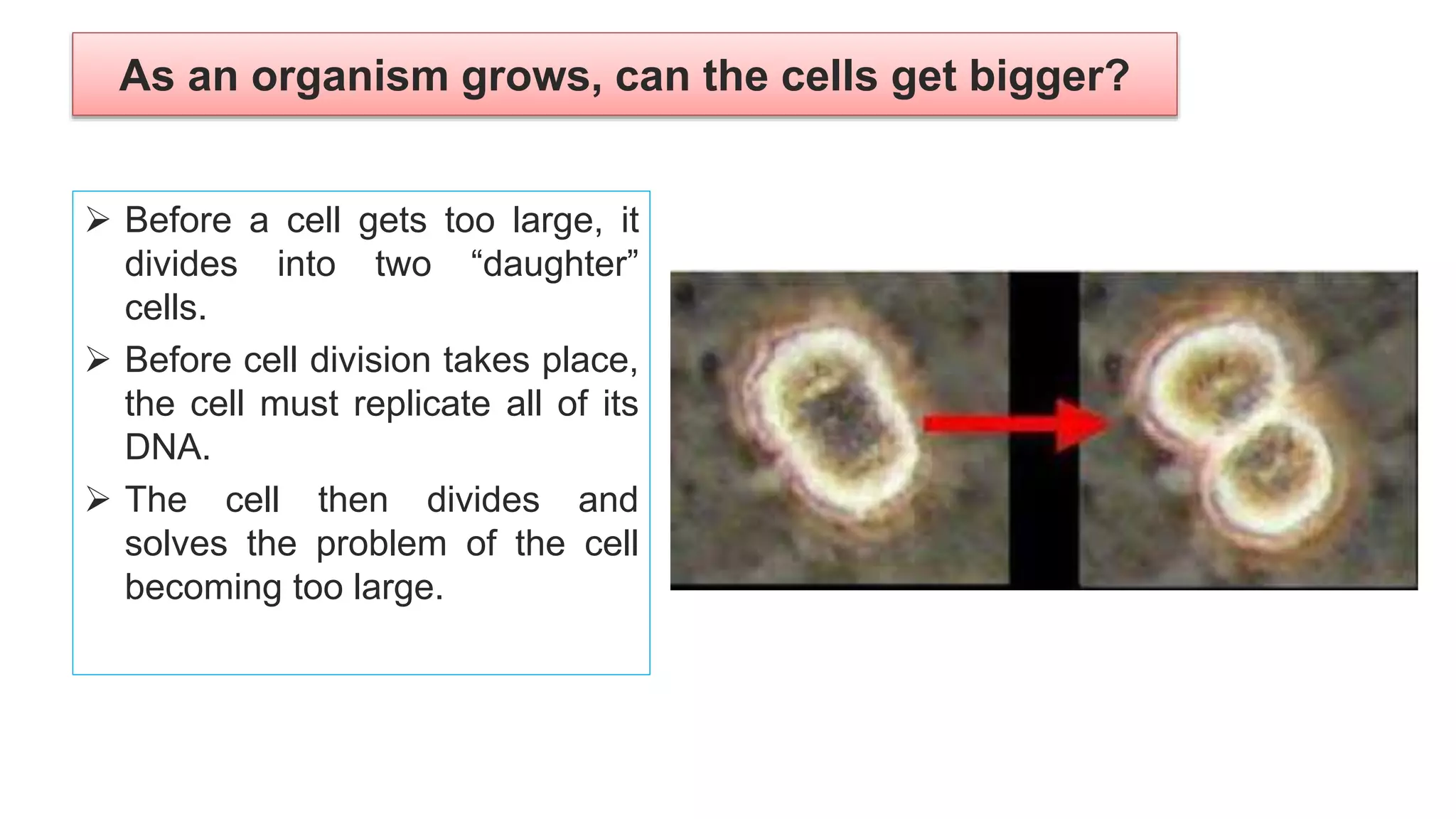
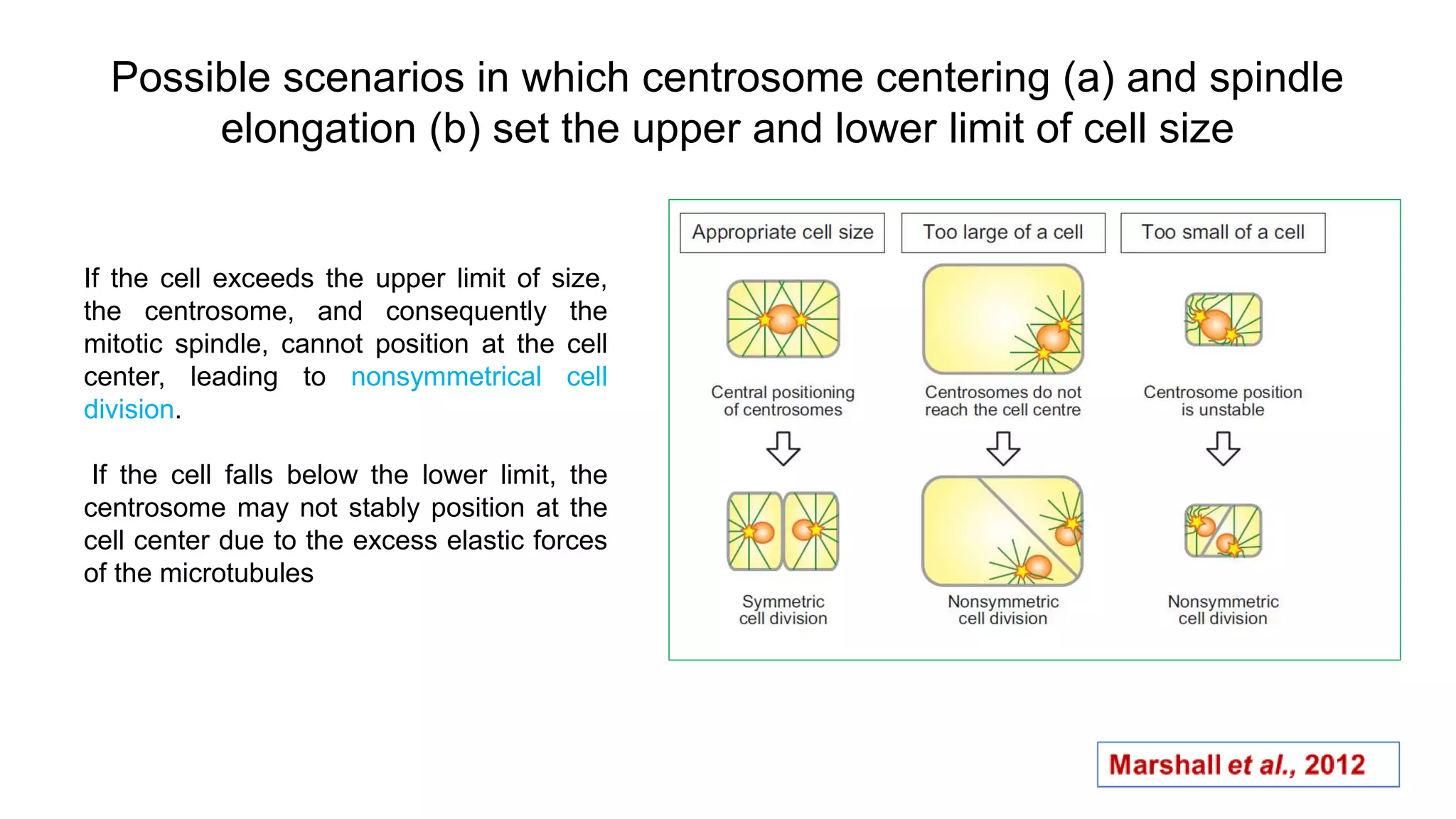
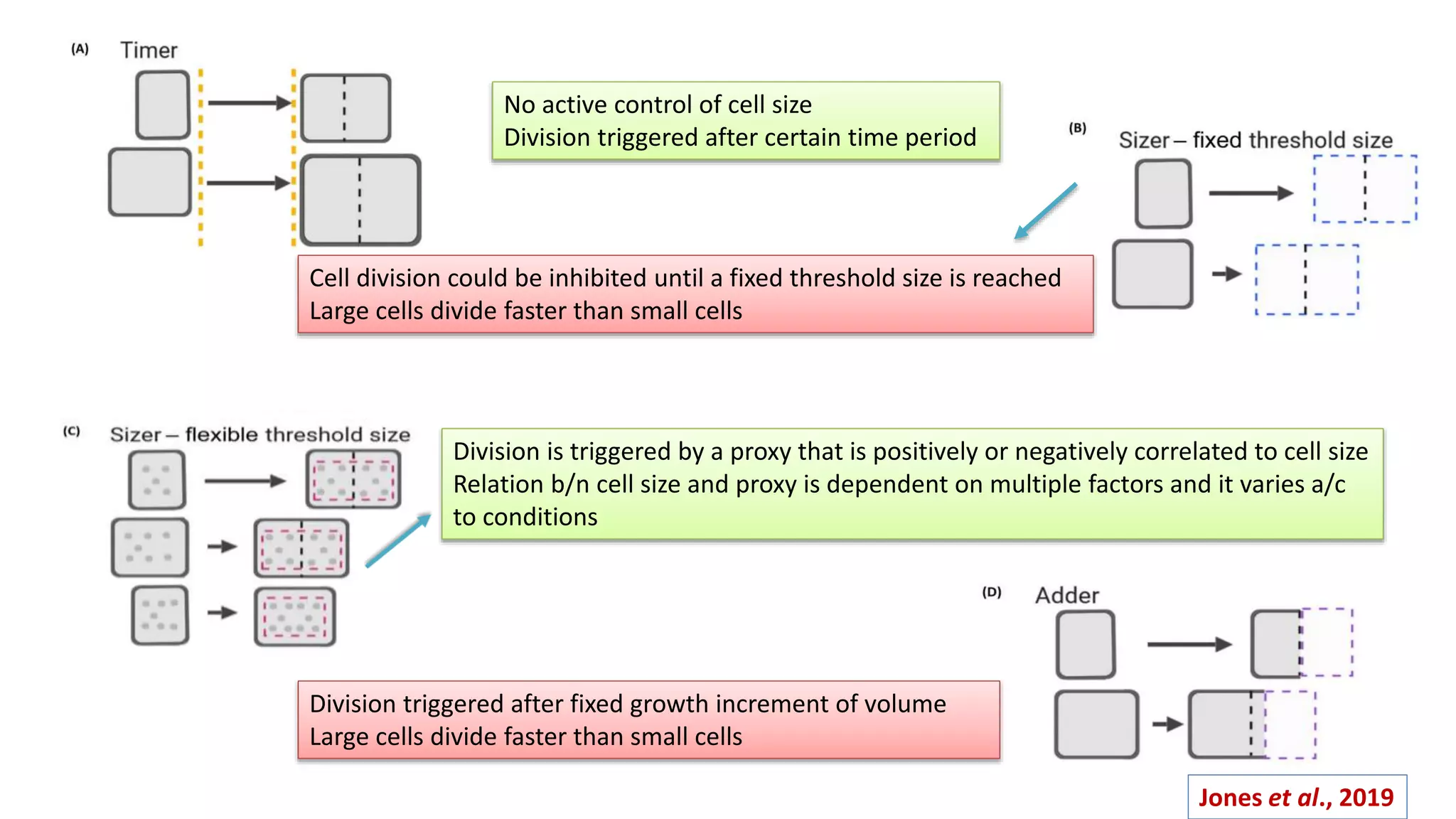
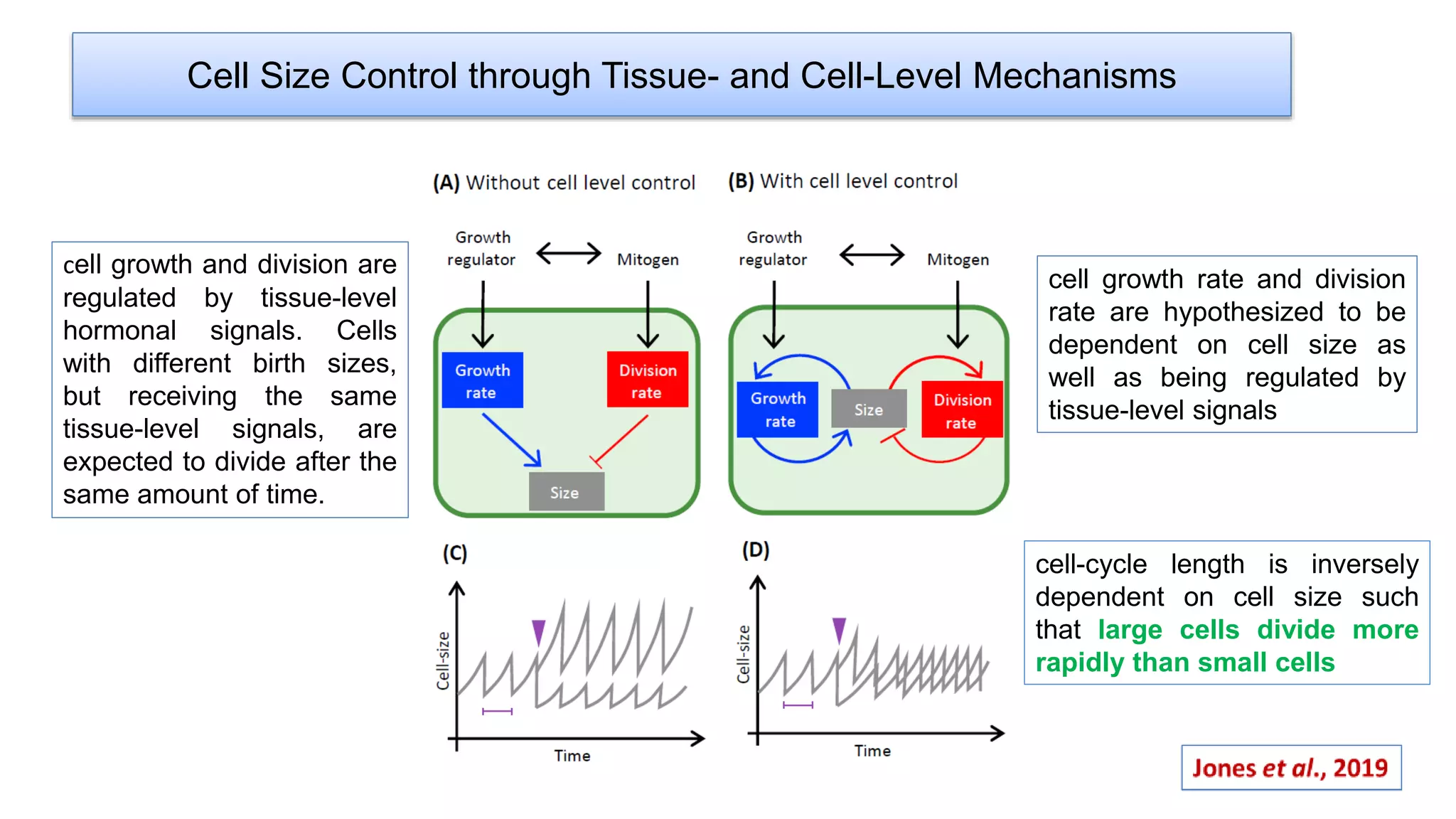
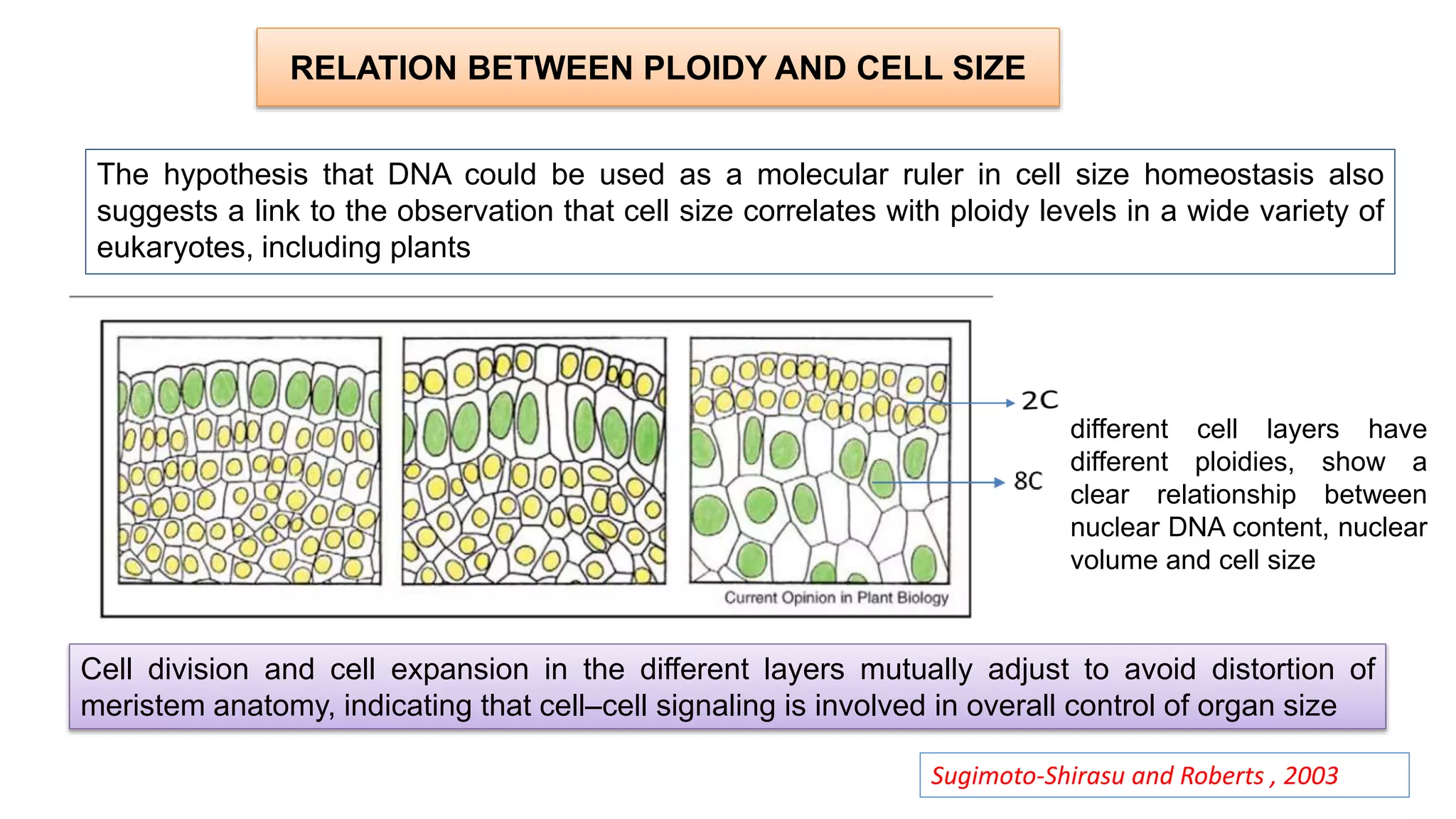
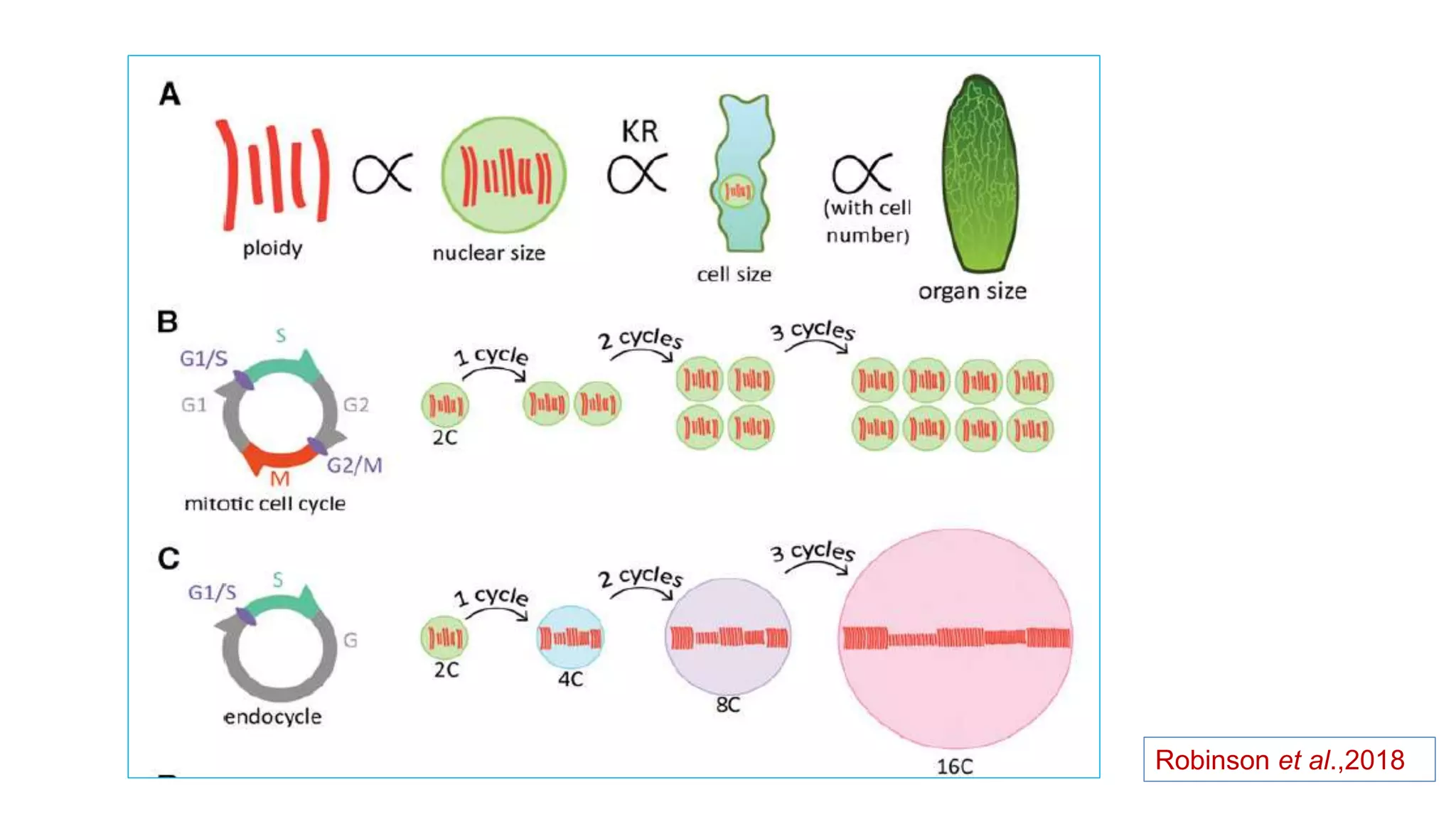
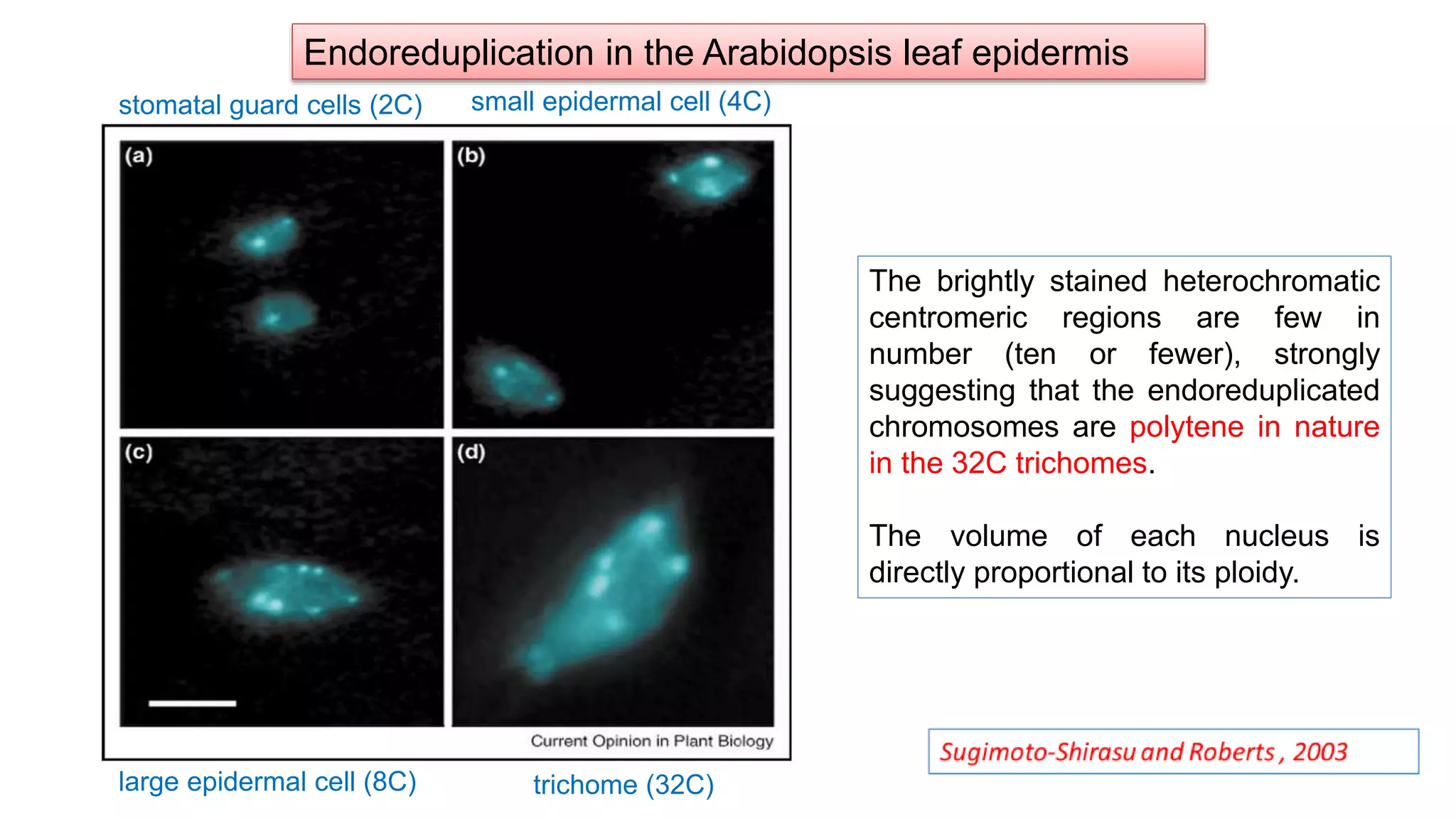
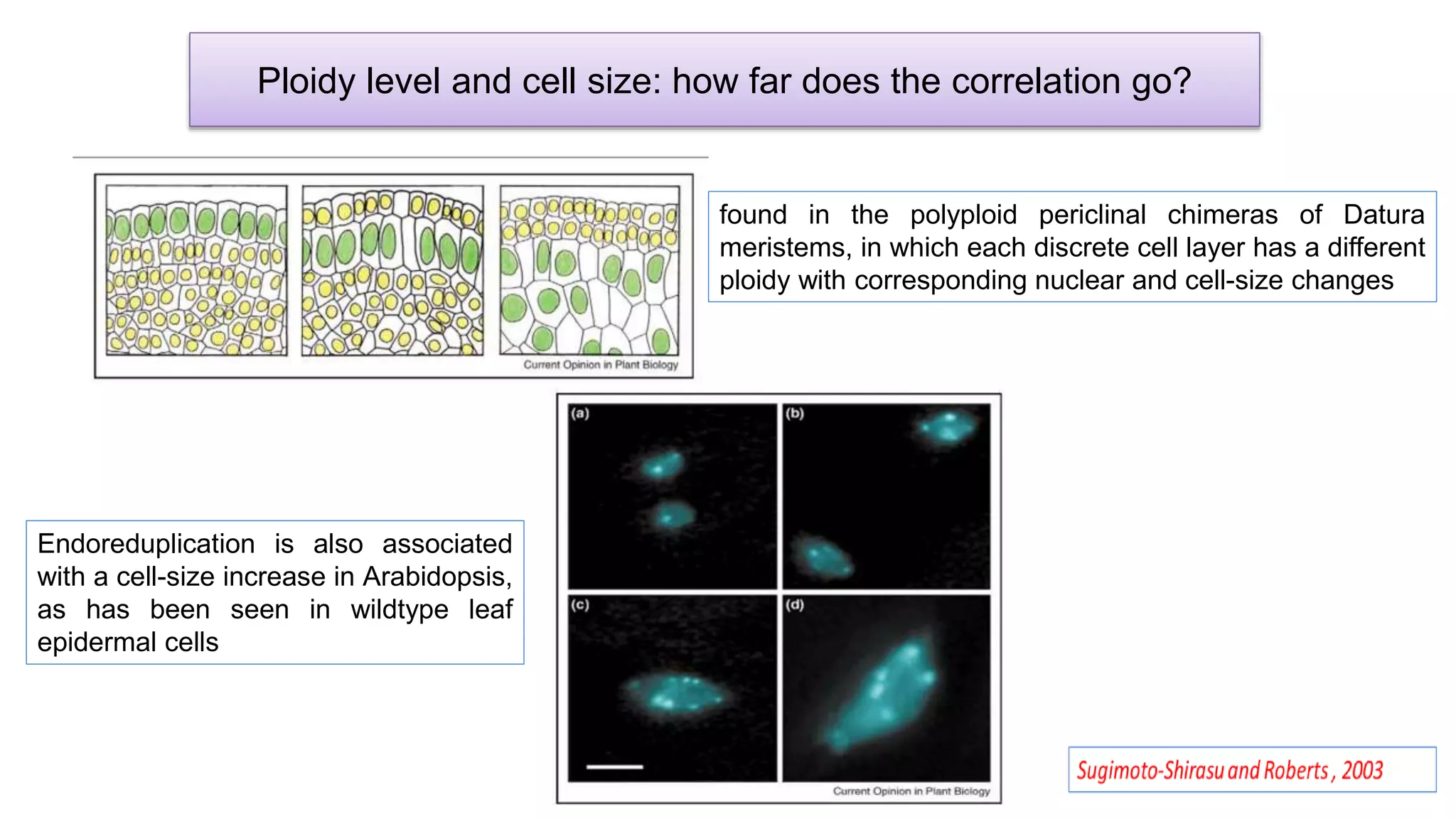
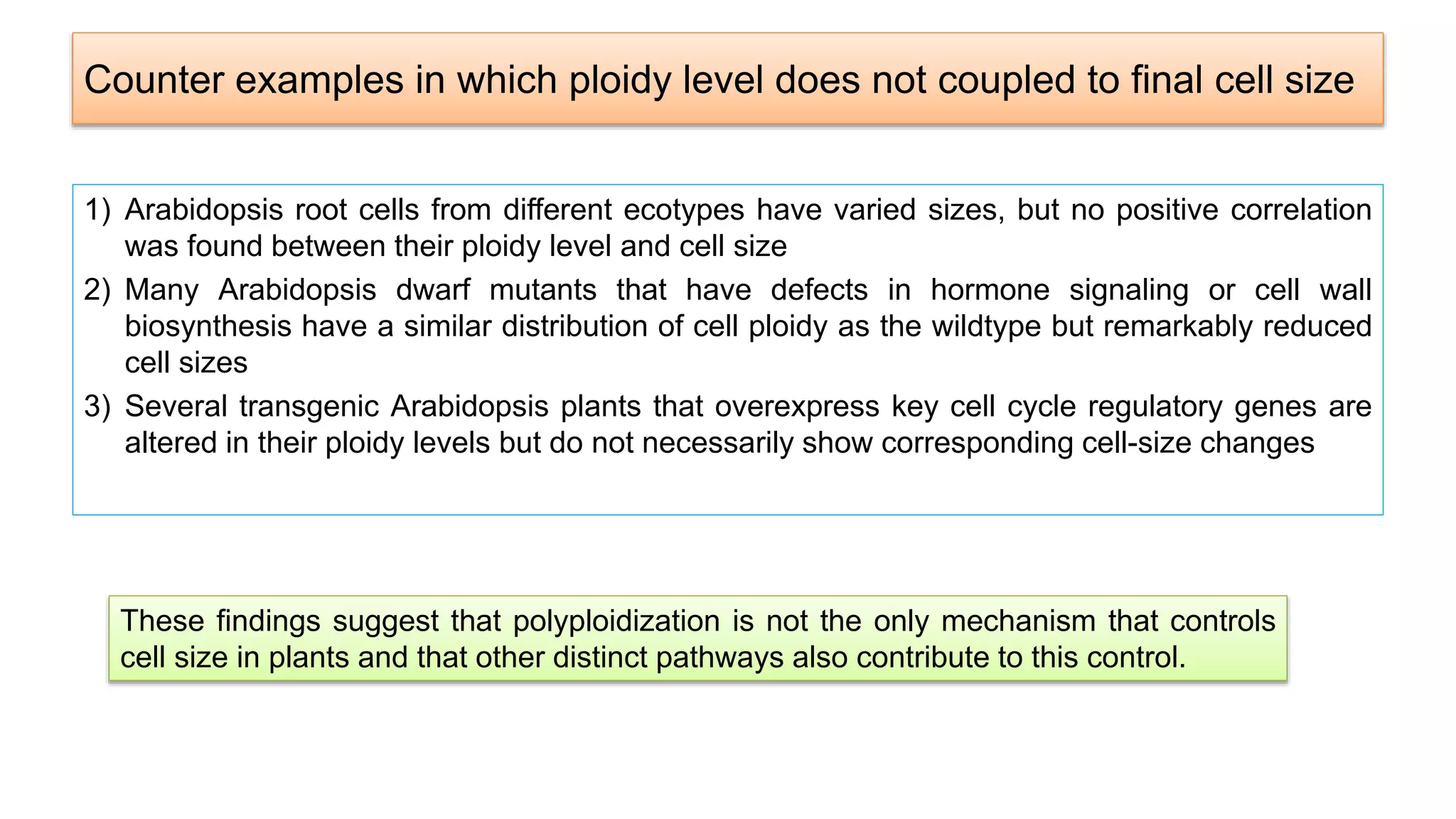
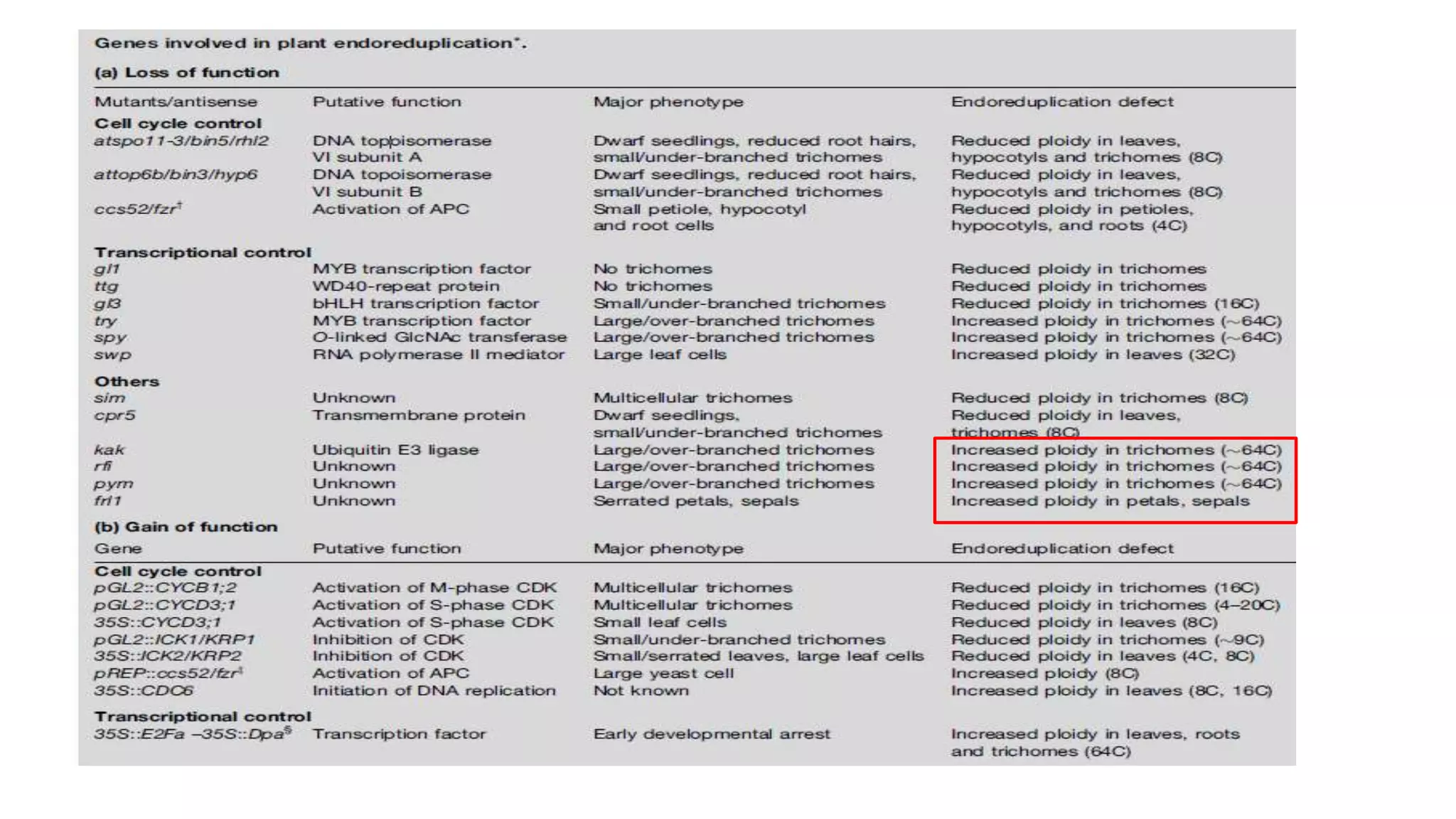
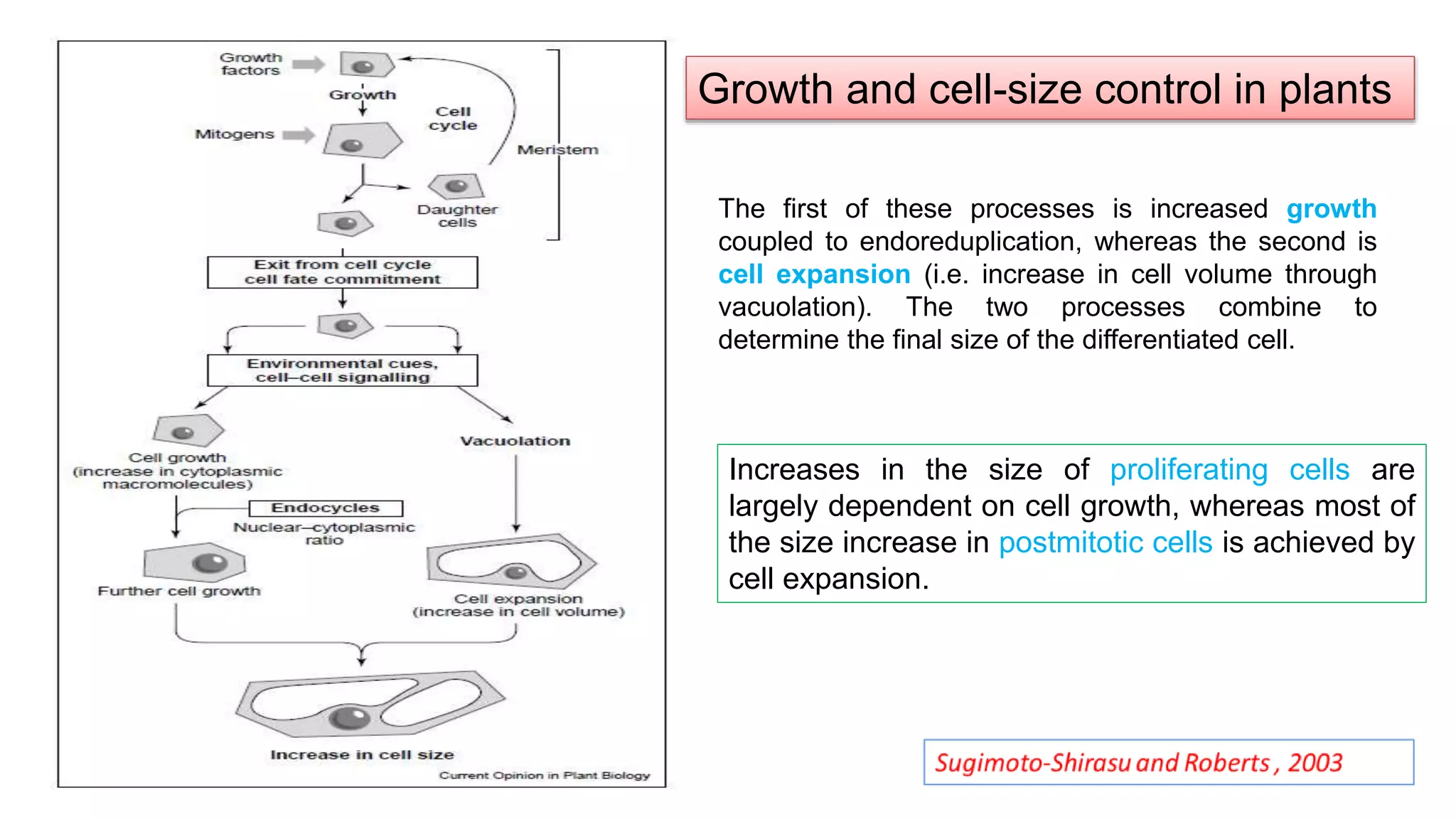
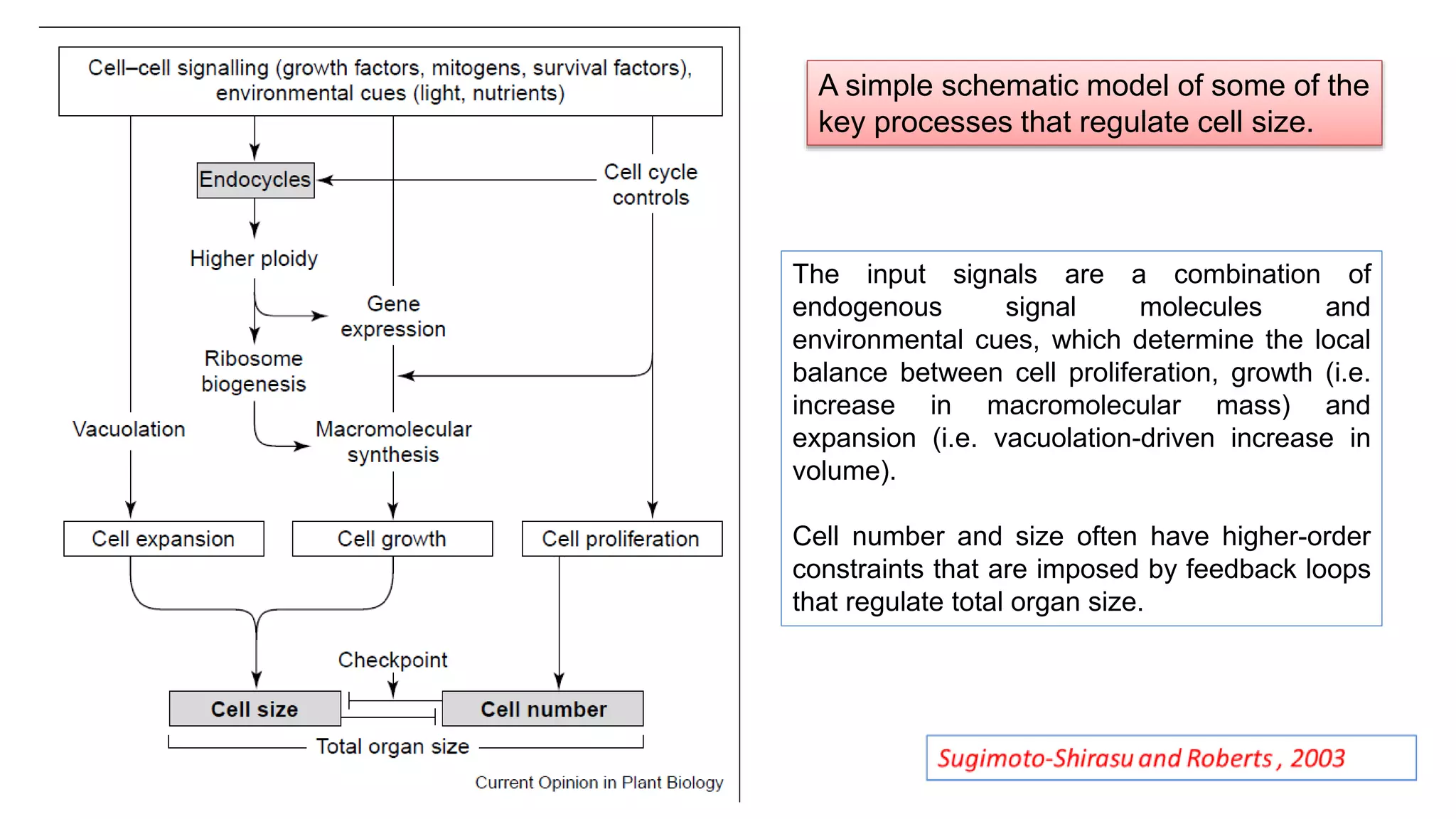
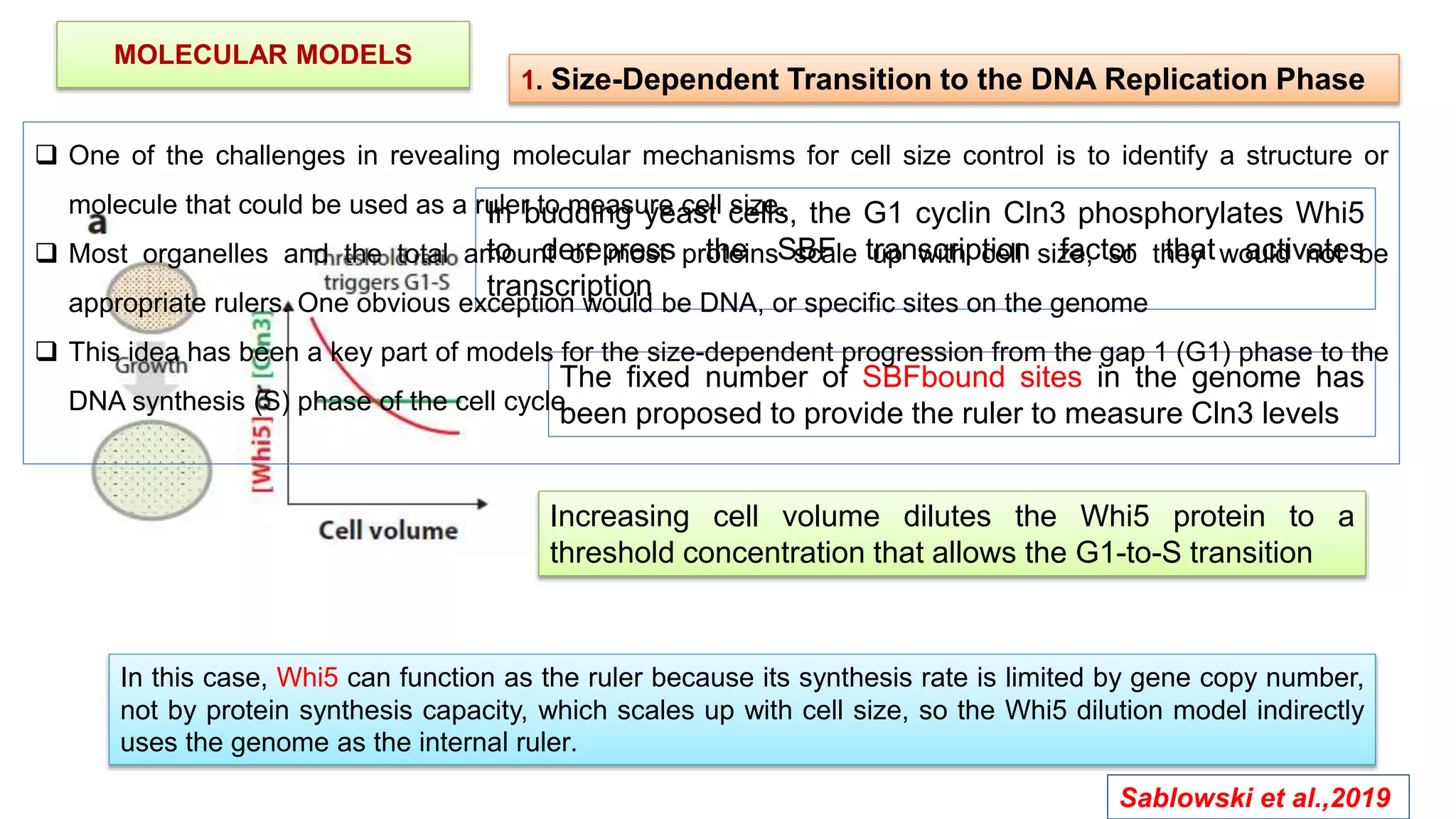
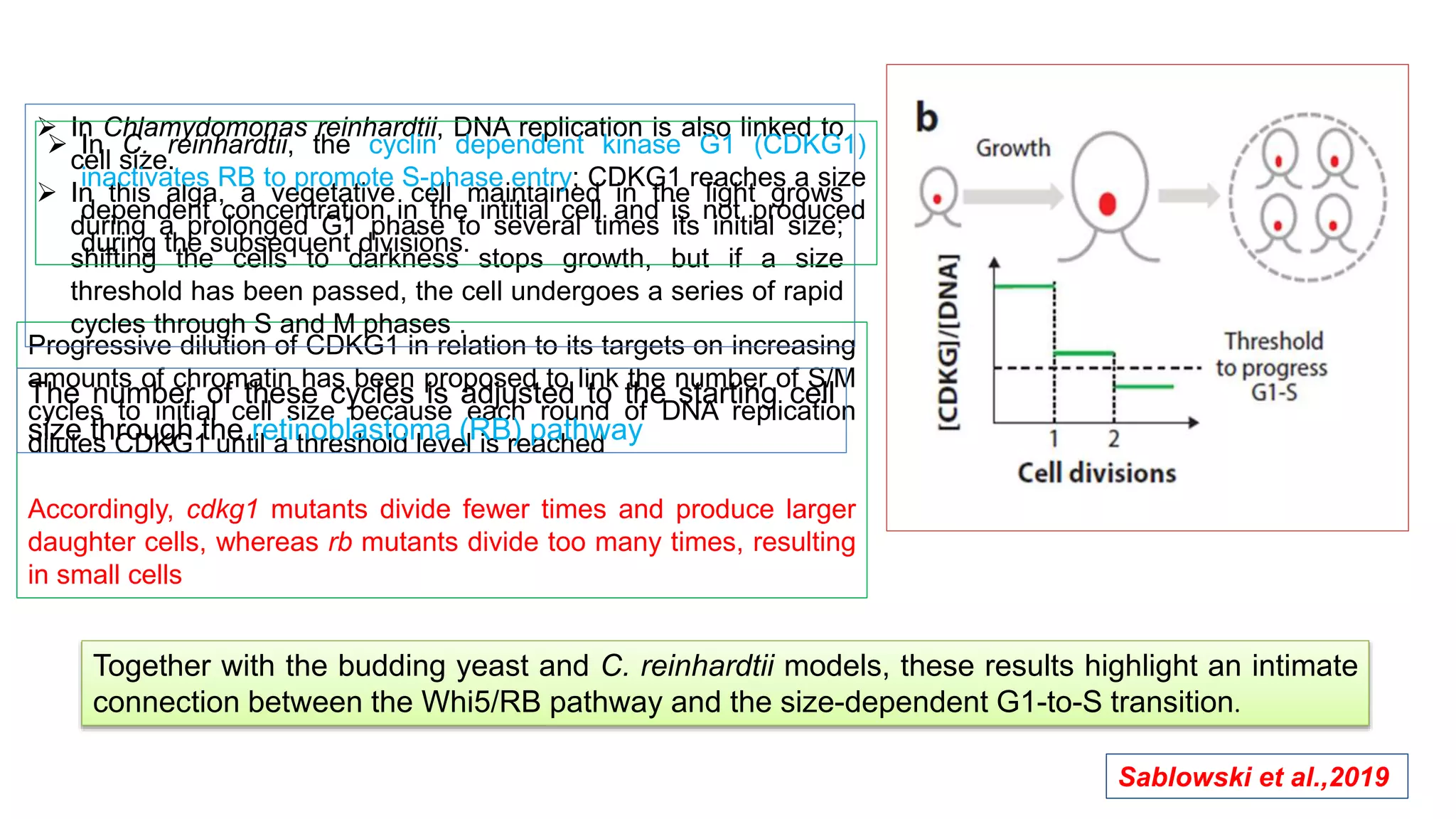
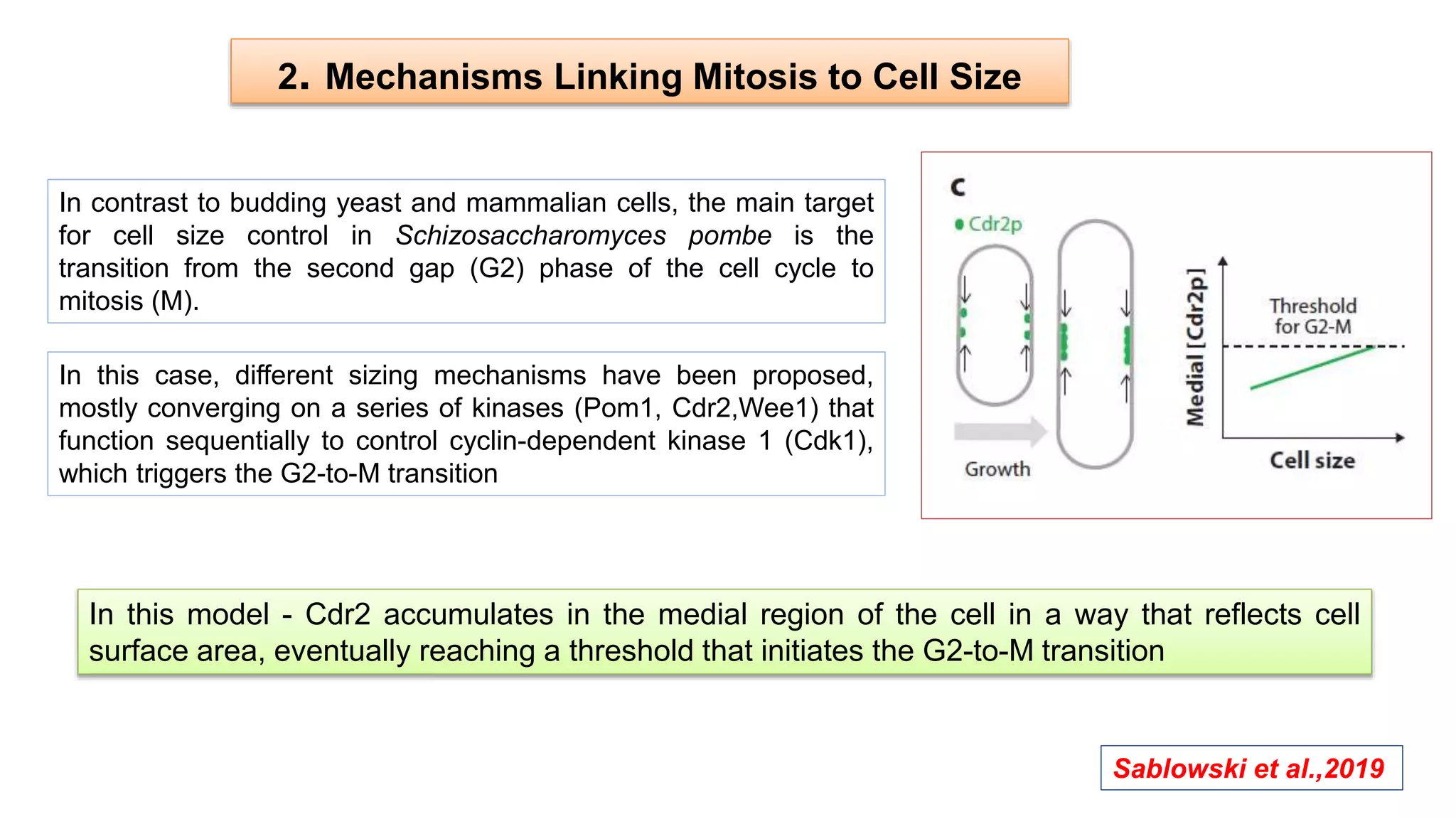
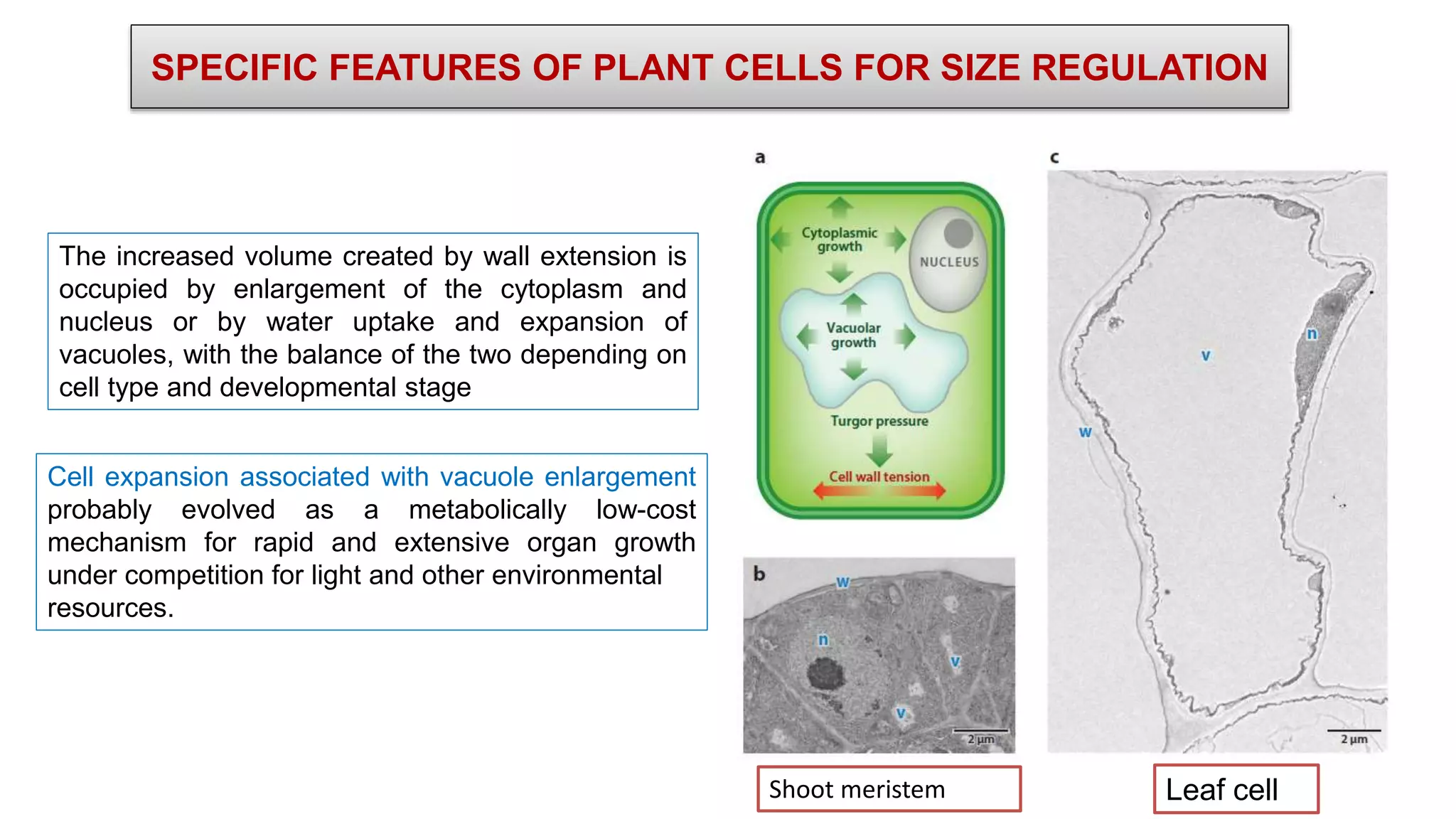
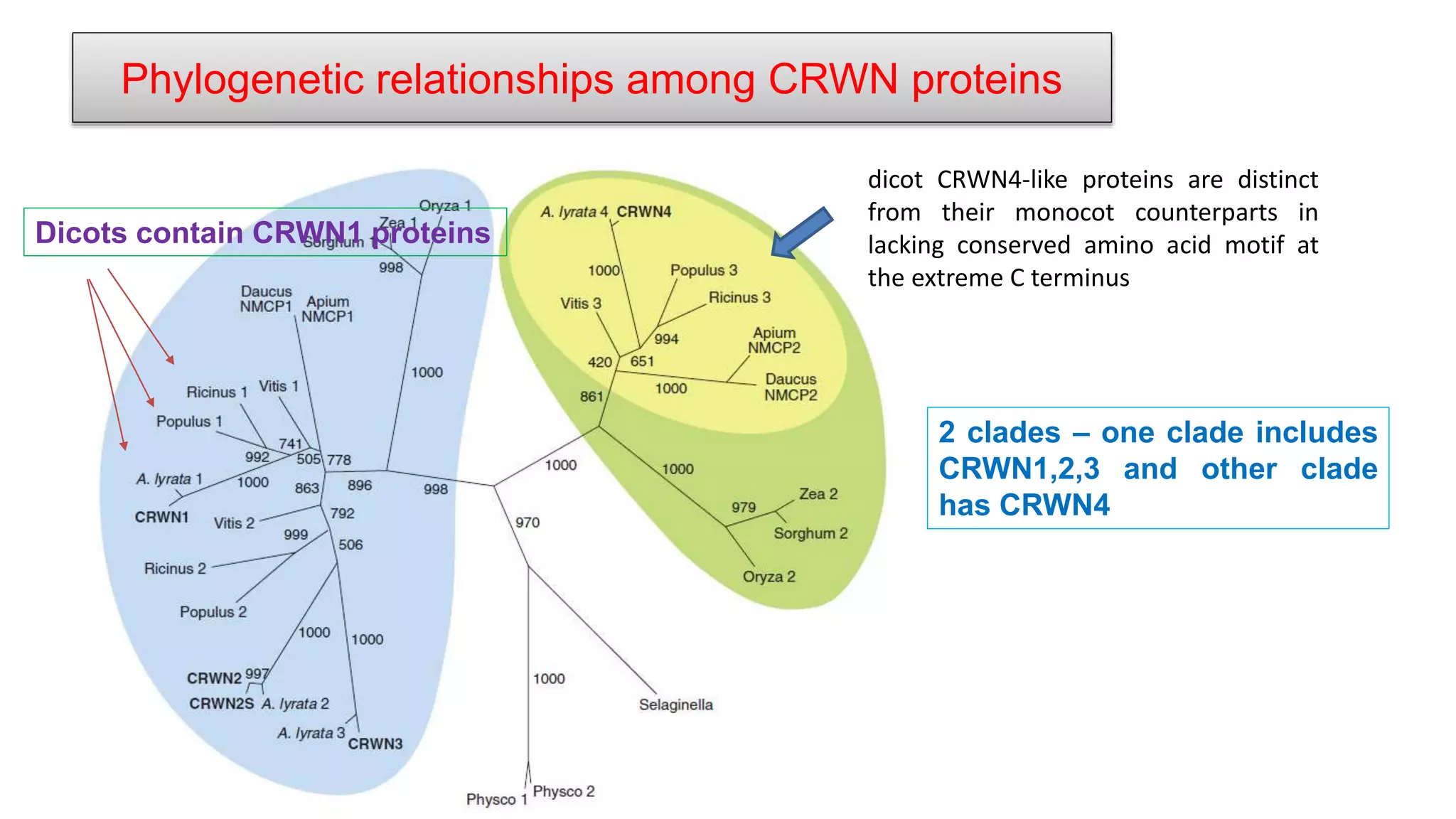
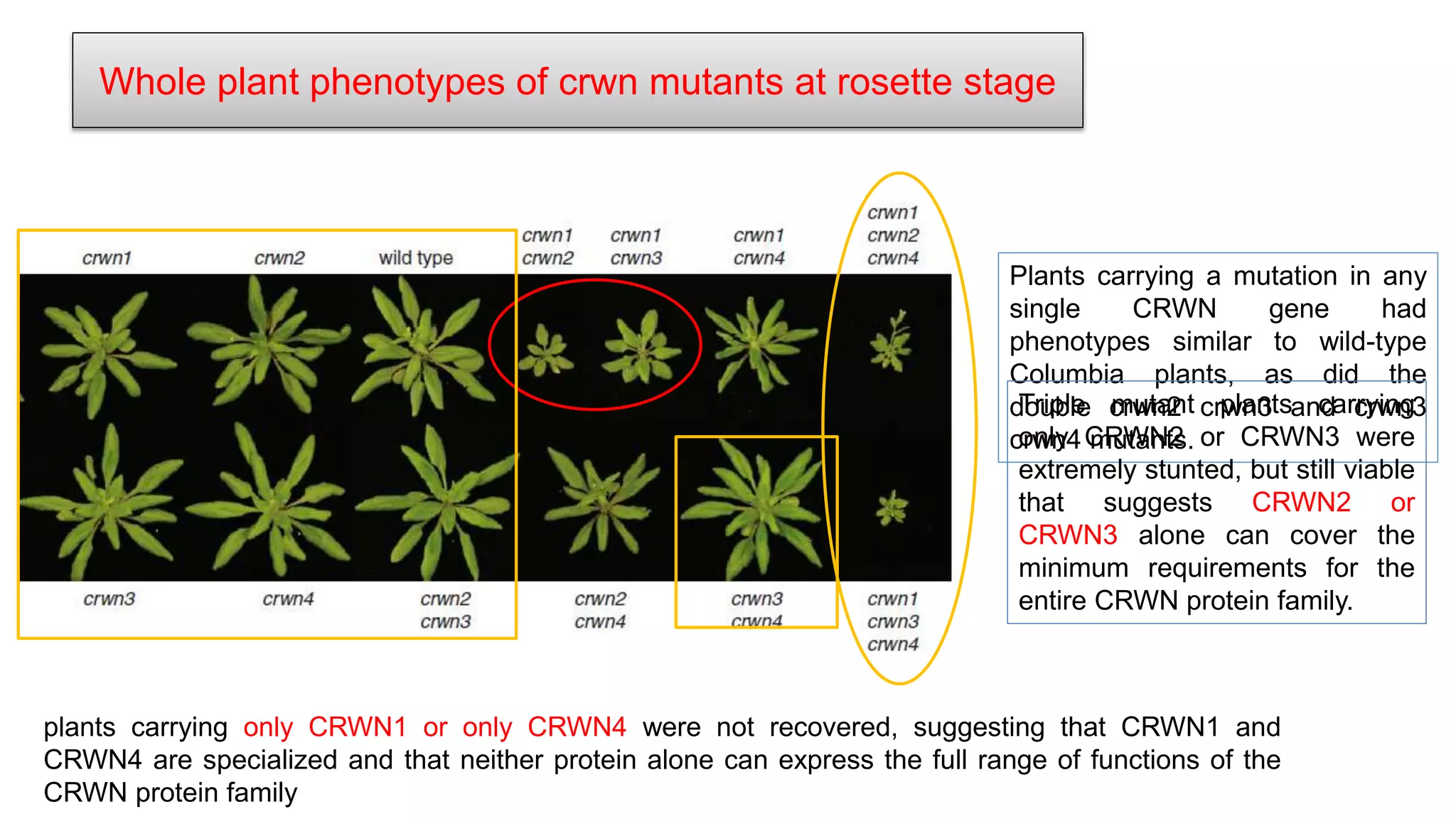
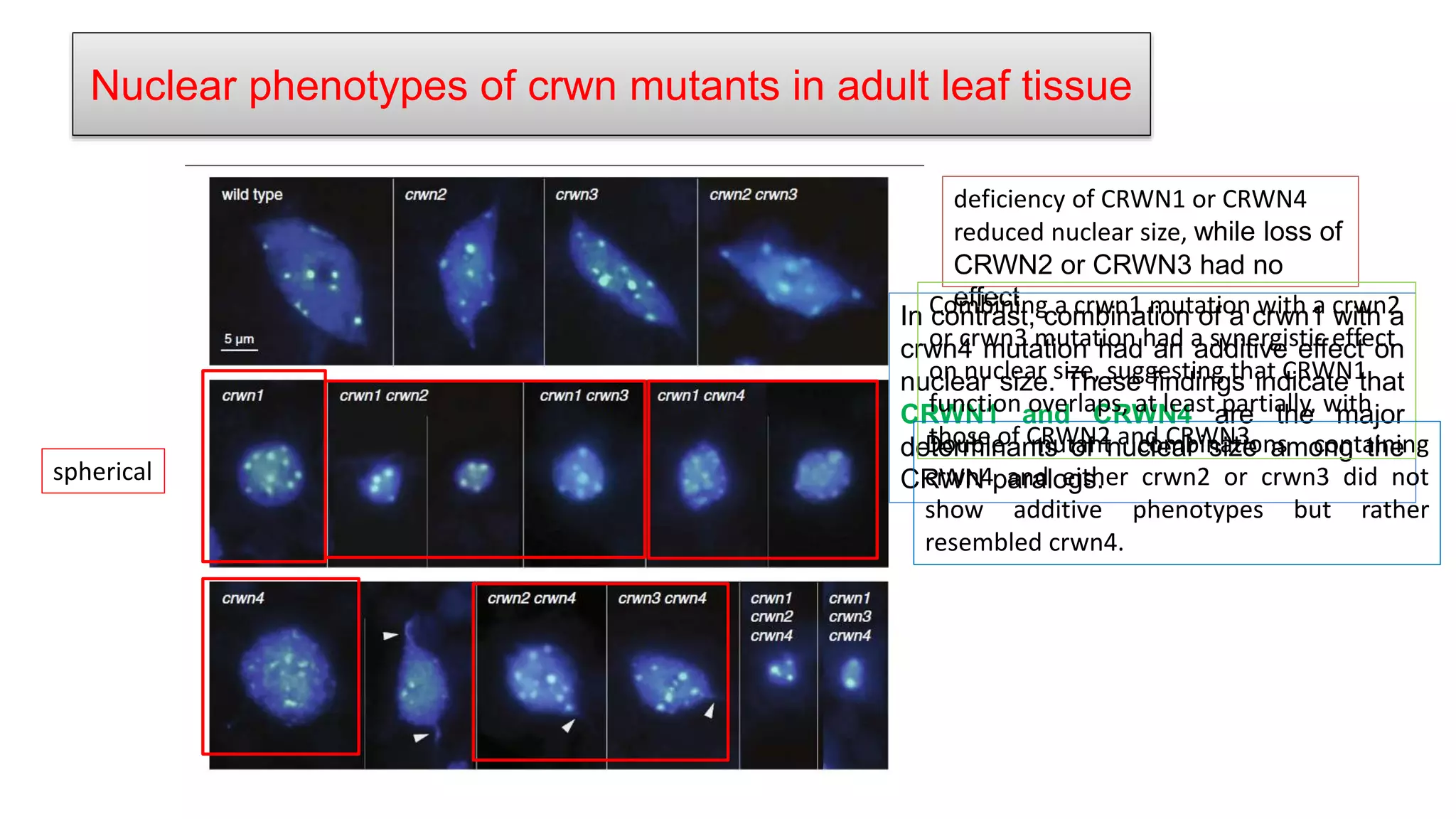
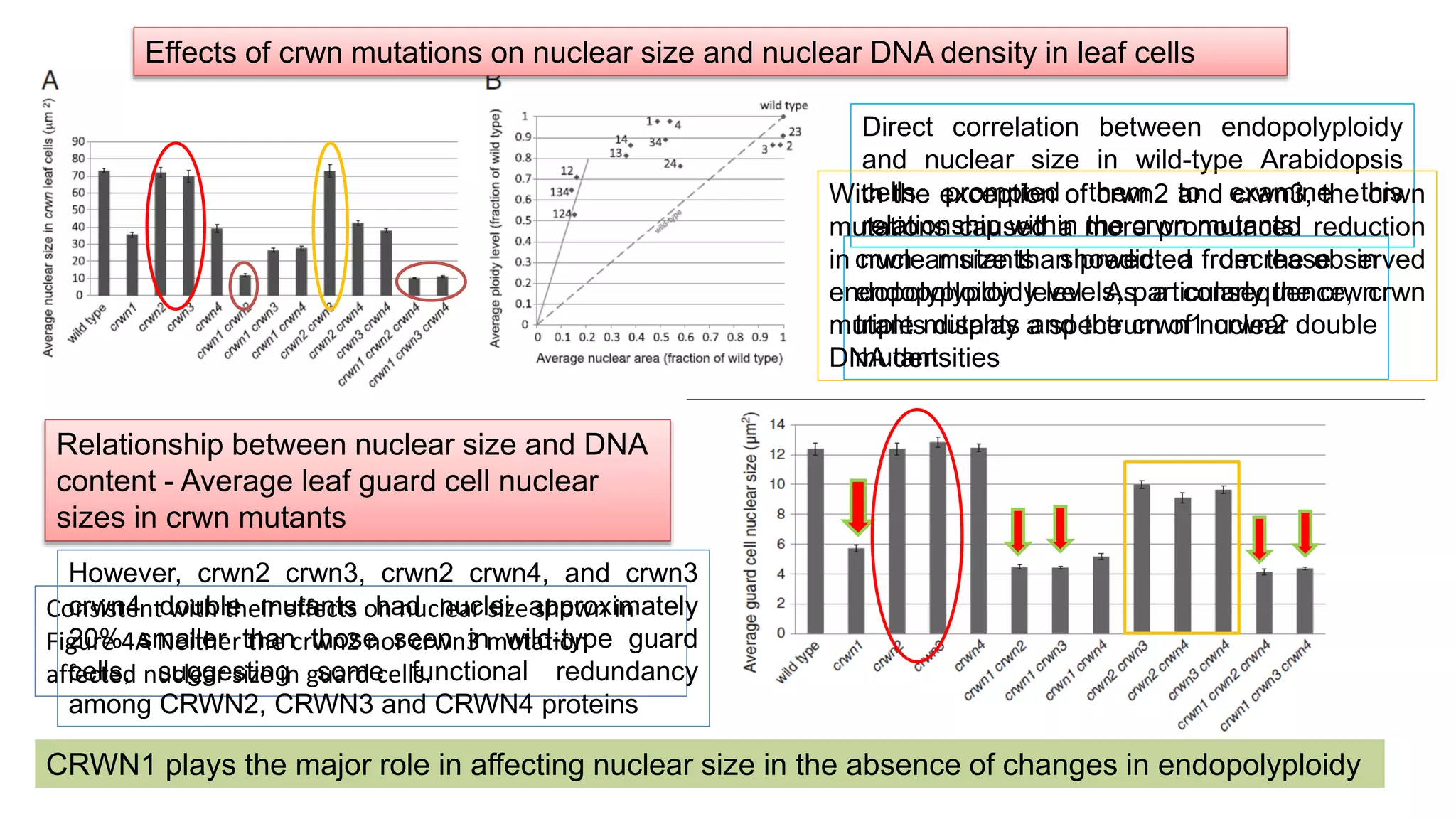
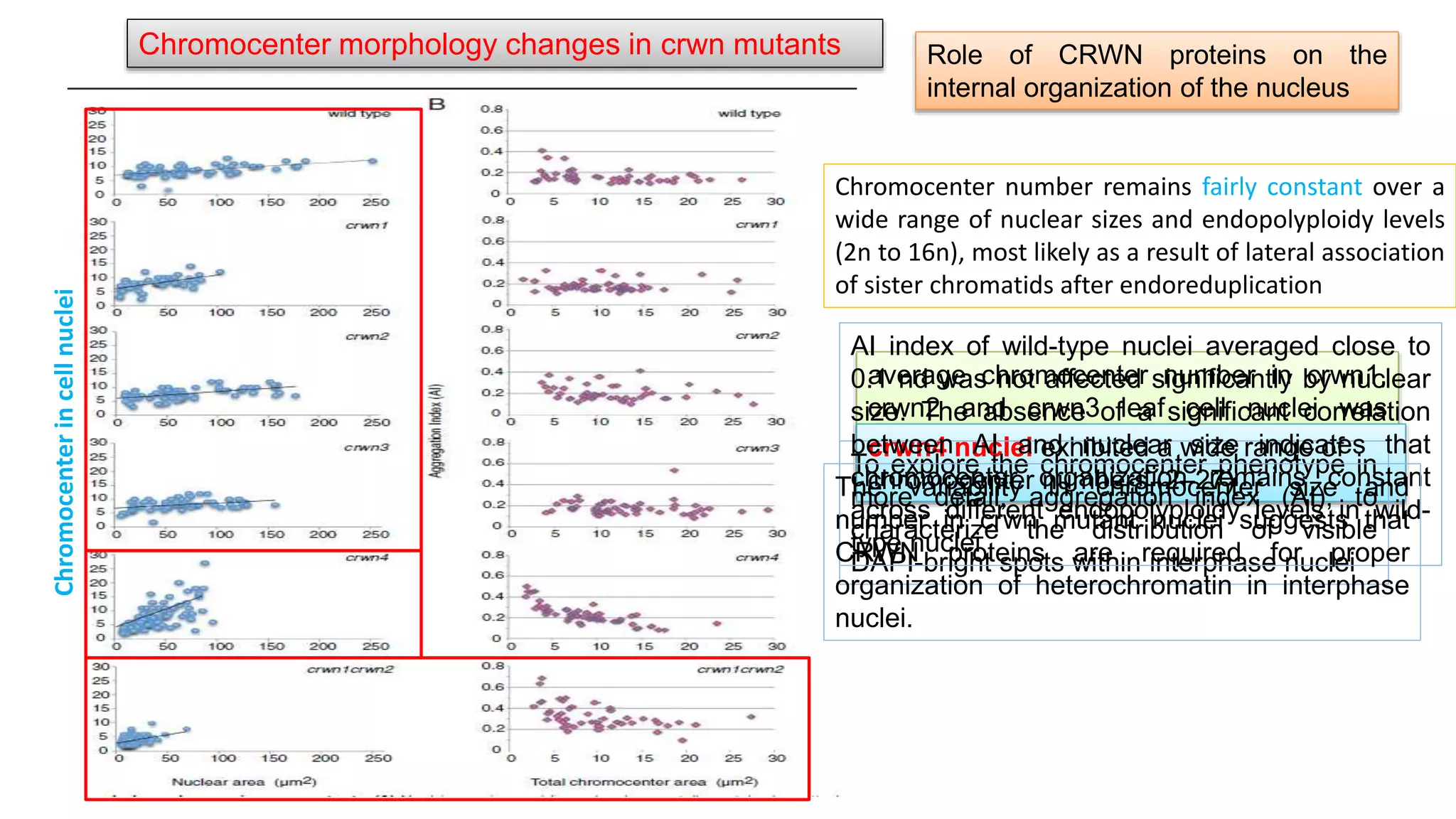
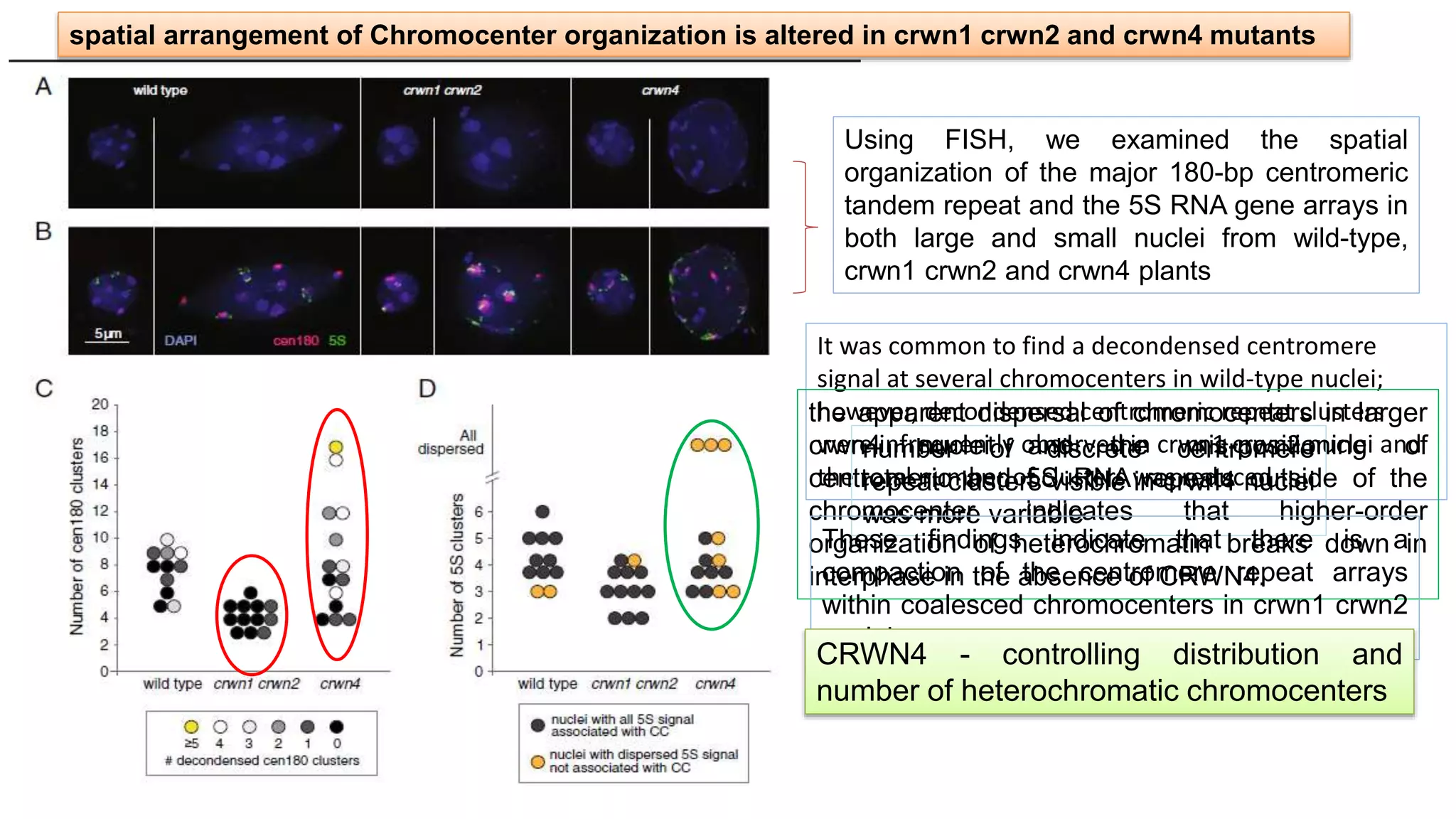
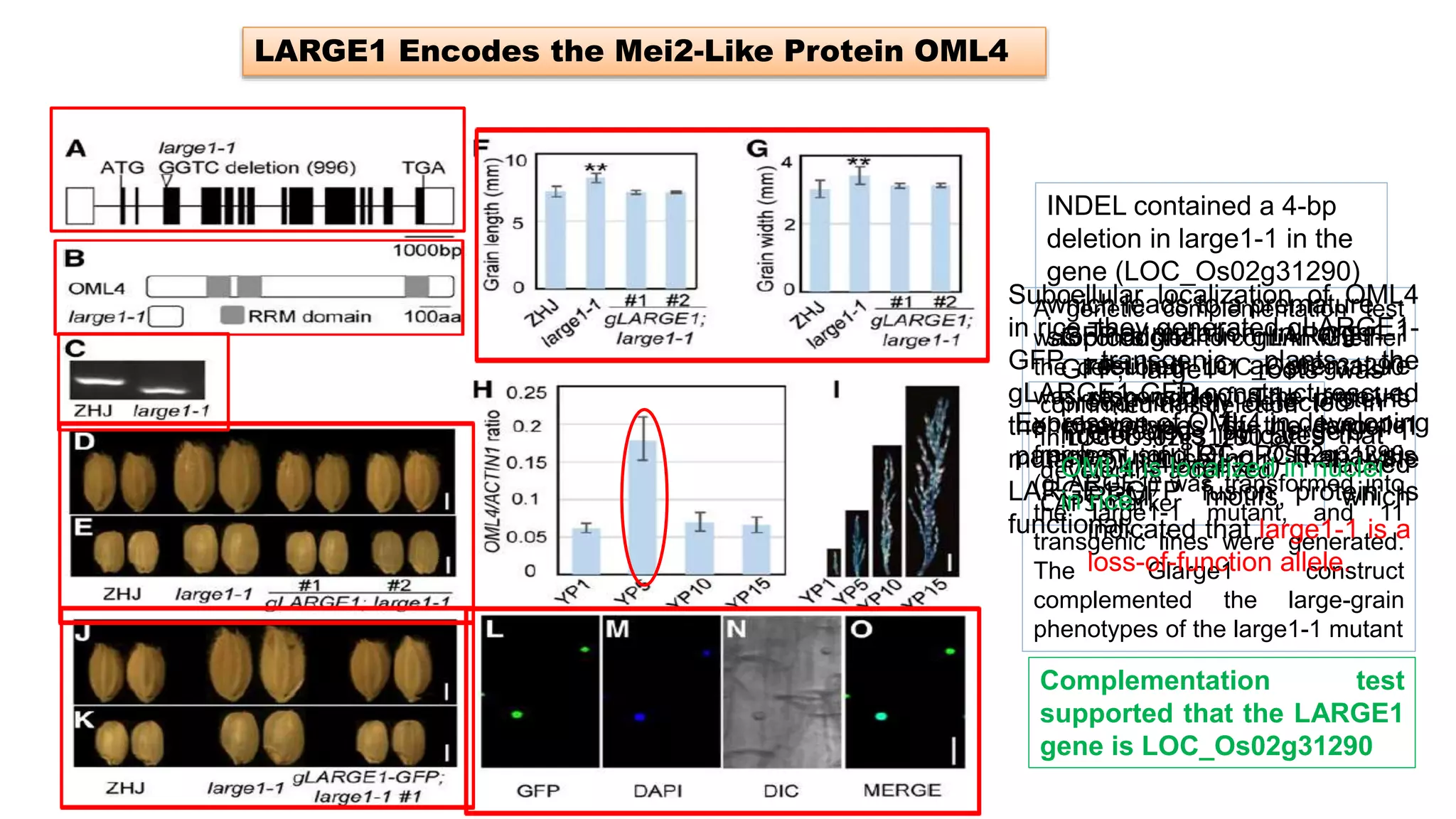
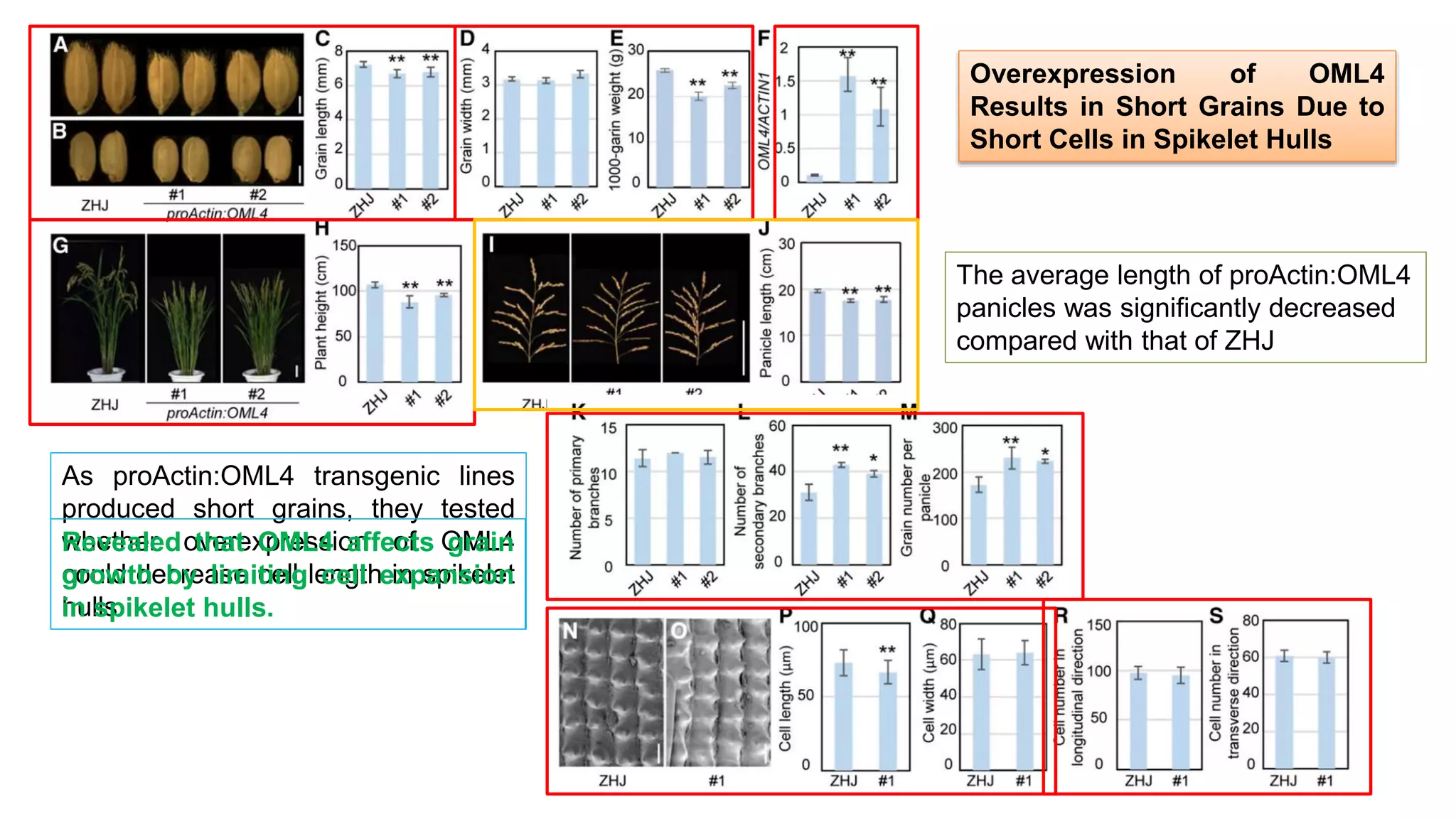
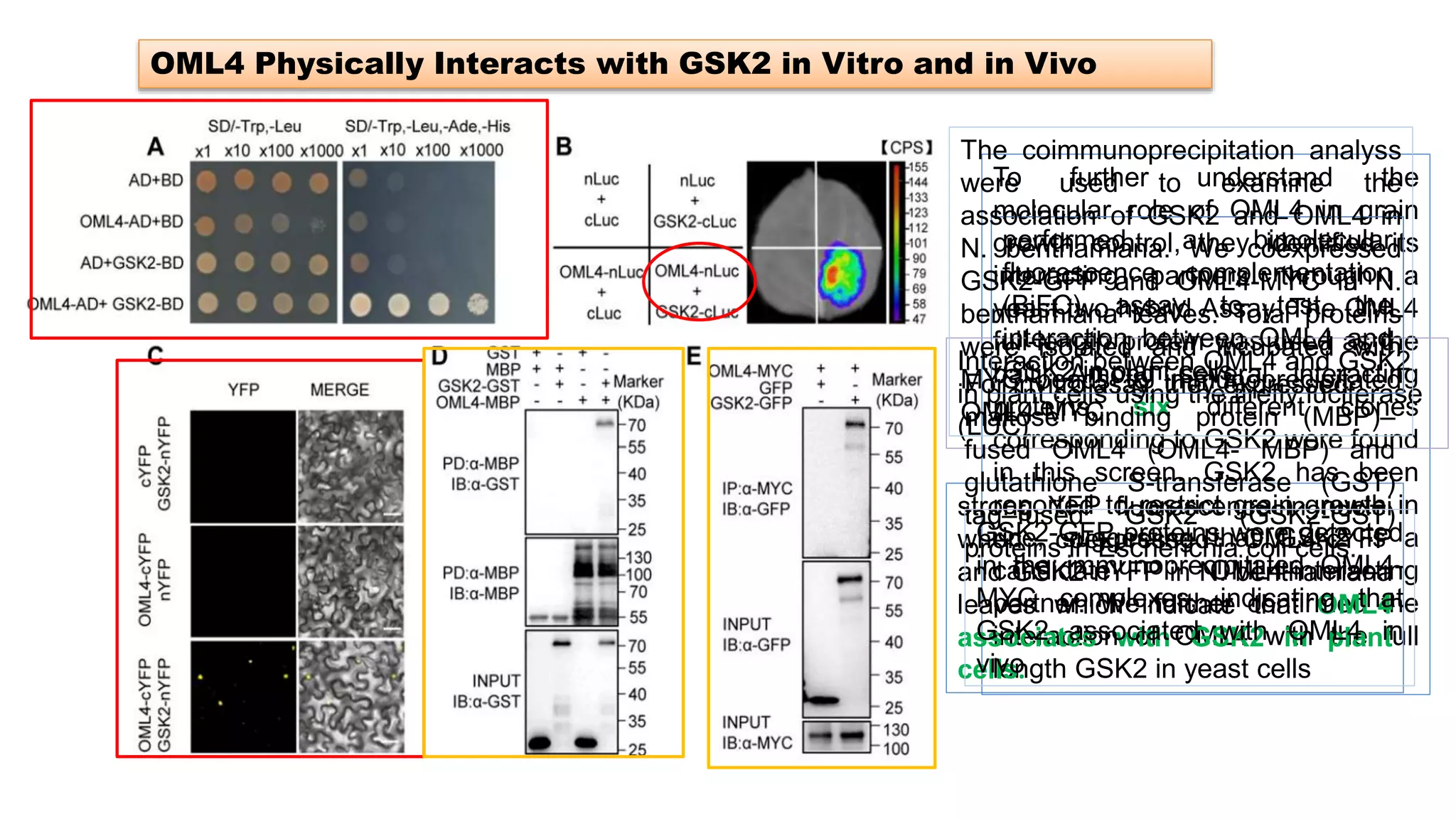
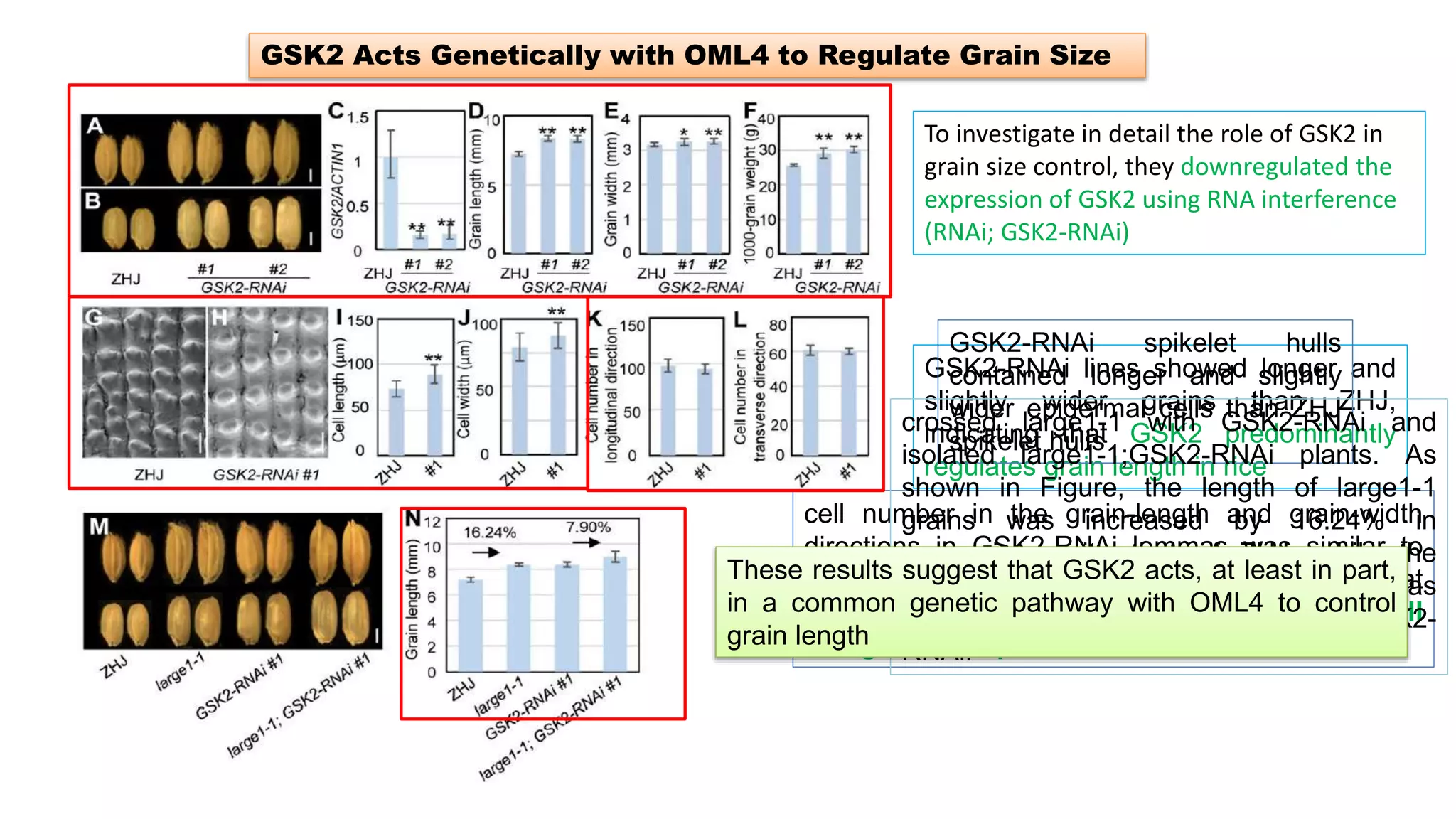
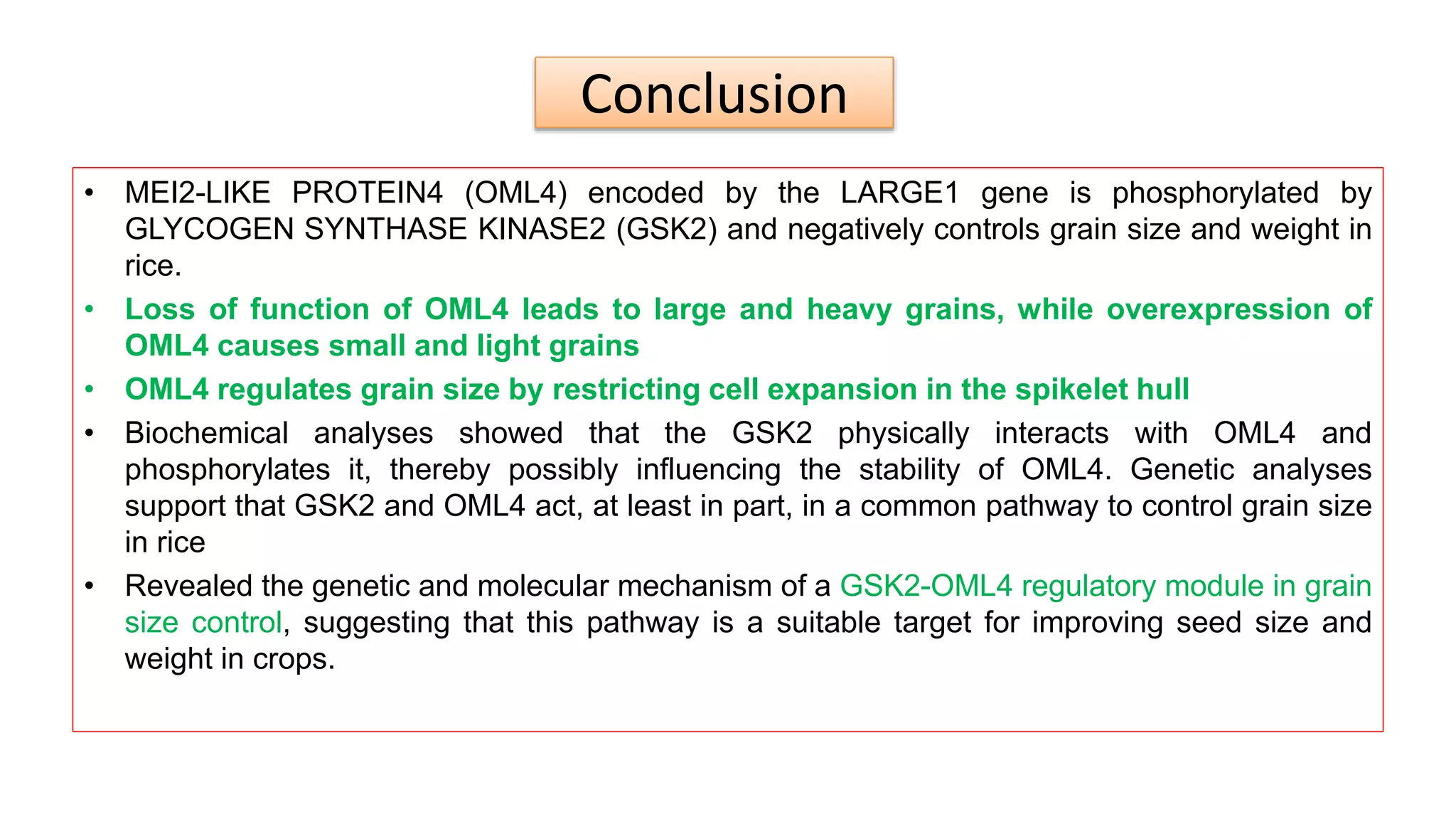
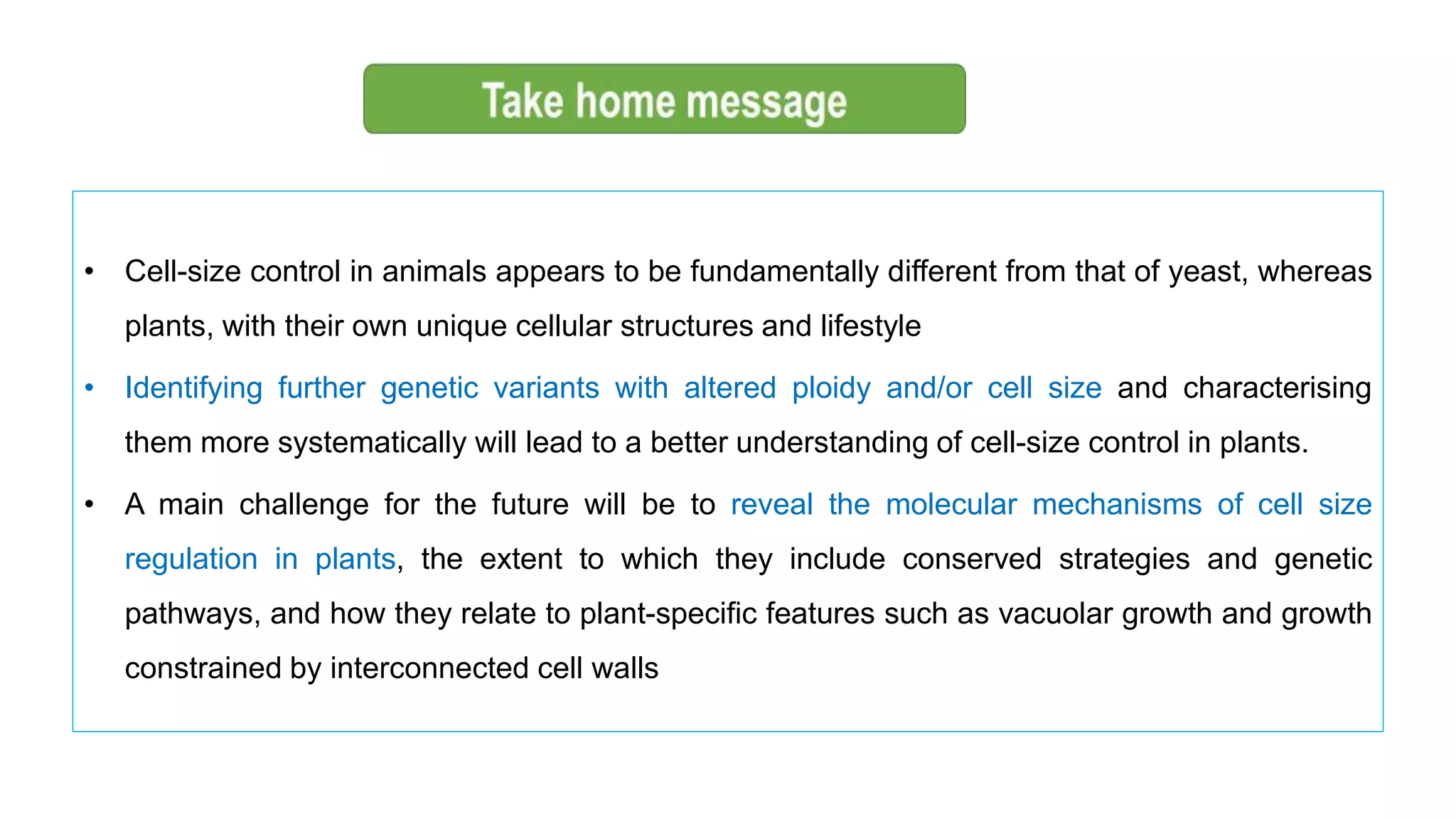
The seminar discusses the mechanisms controlling cell size in plants, highlighting factors such as diffusion, DNA limitations, and surface area-to-volume ratios that necessitate cell division as organisms grow. It explores how endoreduplication, a process in which plants increase ploidy levels, contributes to cell size increases, and evaluates various models of cell size control based on molecular mechanisms and hormonal signals. Additionally, it addresses the role of specific proteins in nuclear size regulation and the complex interplay of size-sensing mechanisms in multicellular plant growth.