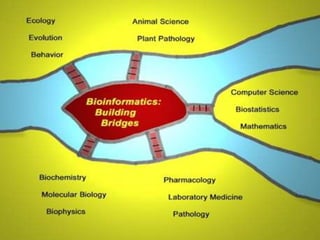
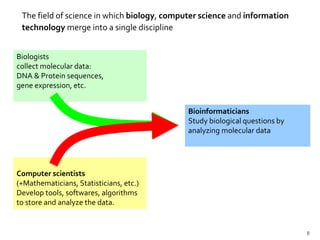
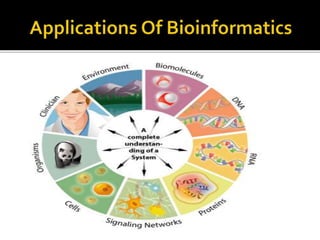
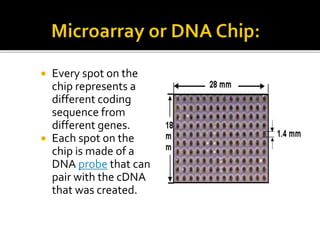
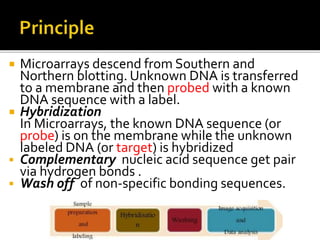
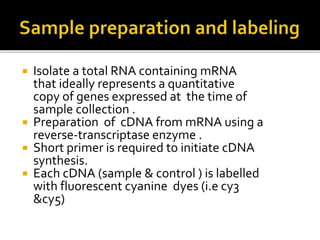
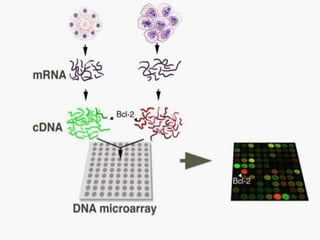
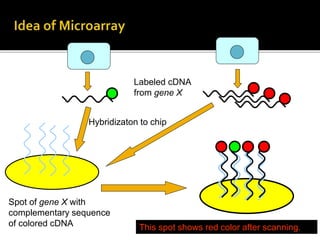
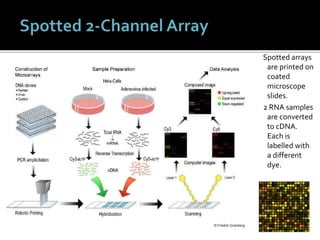
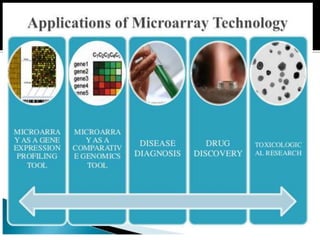
This document provides an overview of the field of bioinformatics, including its history and applications. It discusses how bioinformatics merges biology, computer science, and information technology. It also summarizes key applications like using bioinformatics for human and animal genomics, molecular medicine, microbiology, and more. Microarray technology is introduced, explaining how DNA microarrays work to analyze gene expression levels. Different types of microarrays and platforms are also outlined.