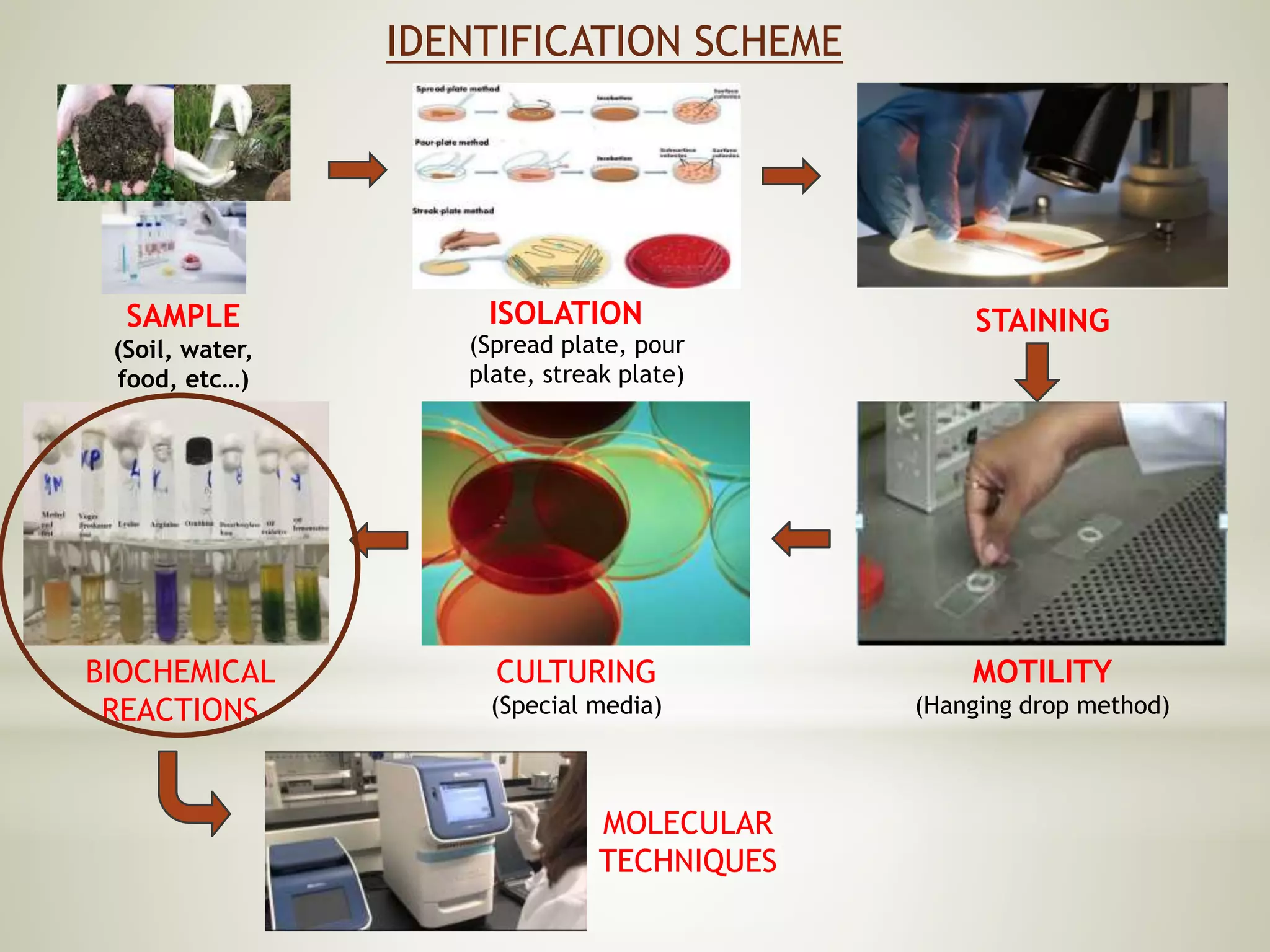
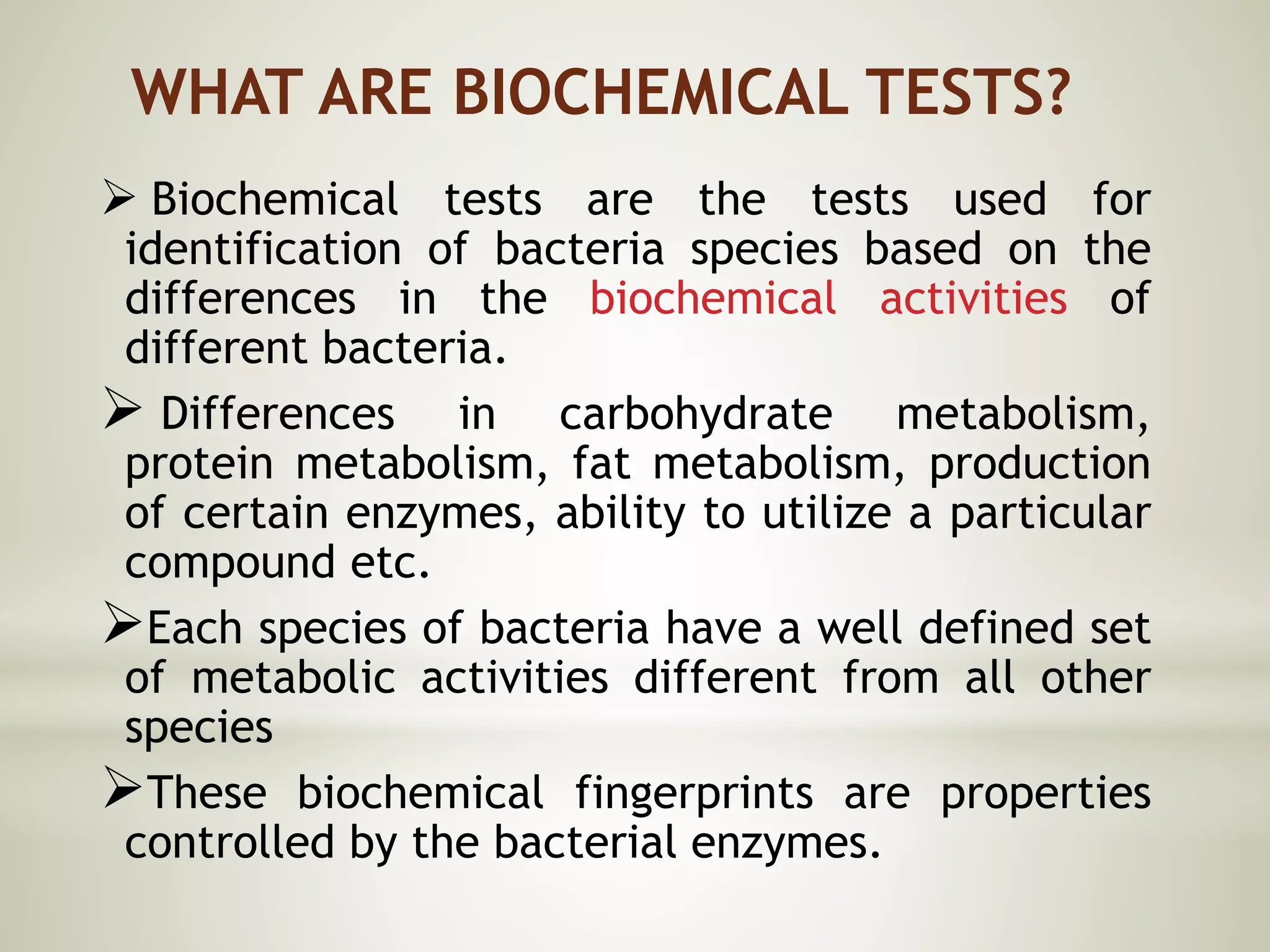
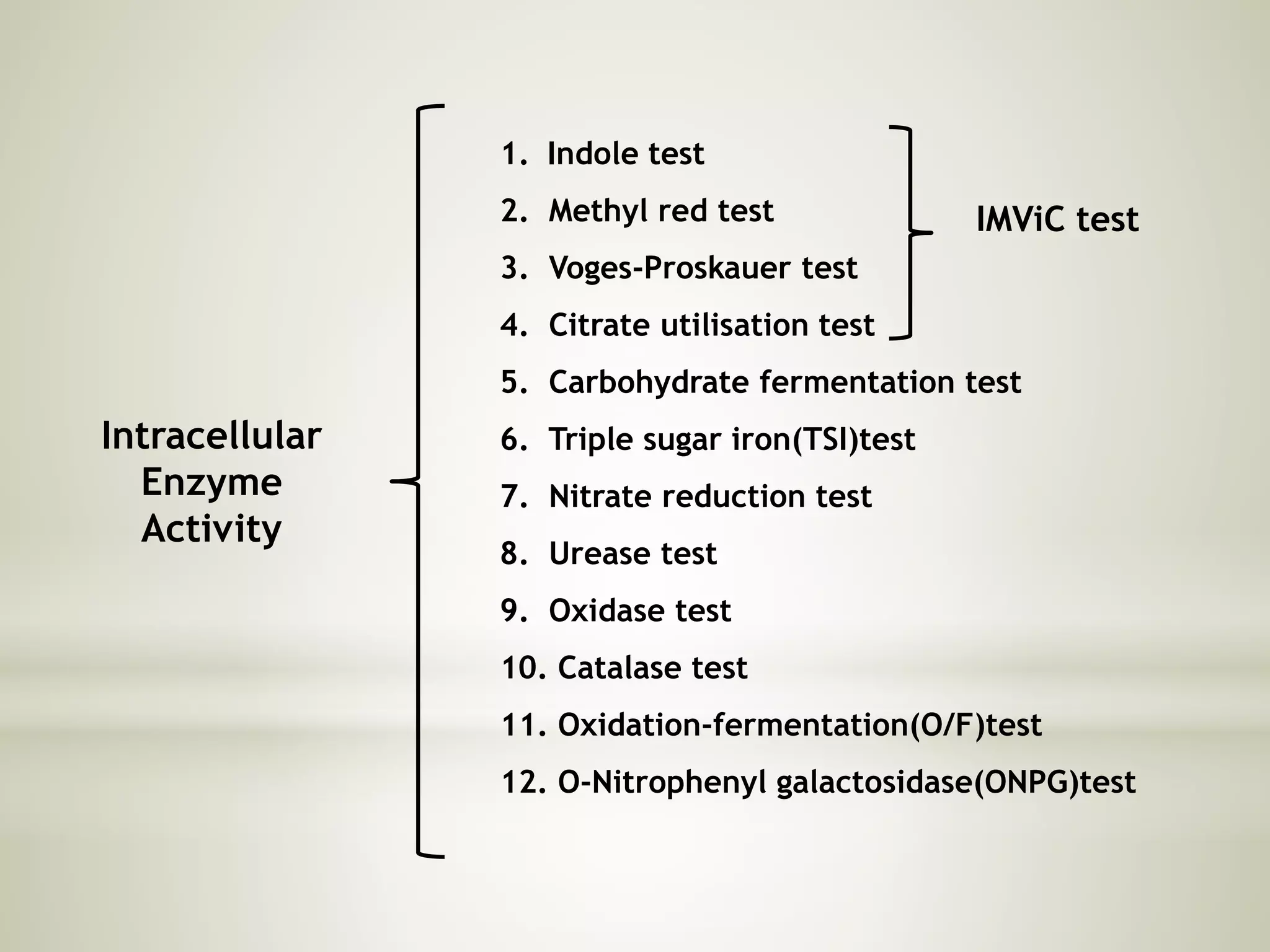
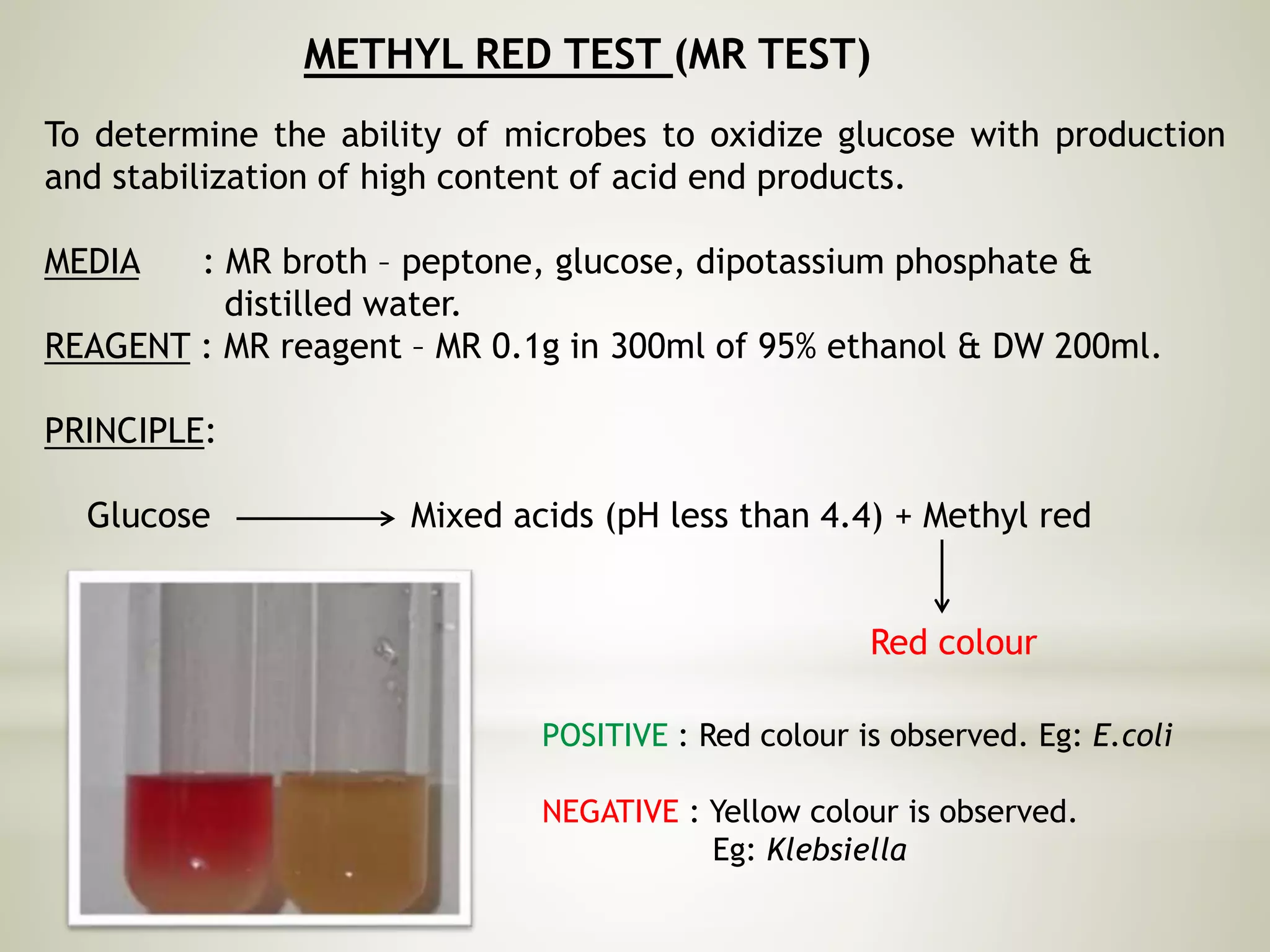
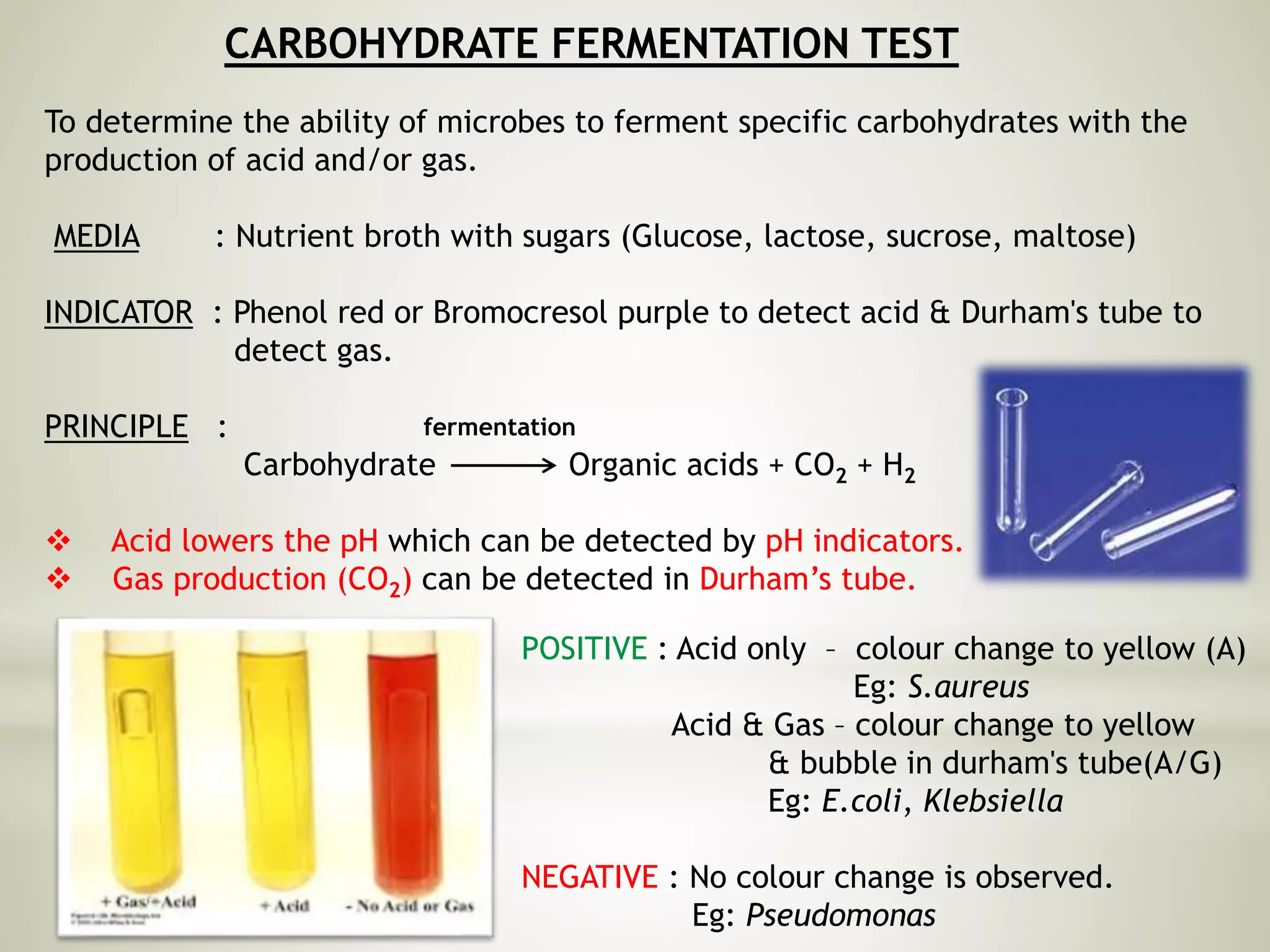
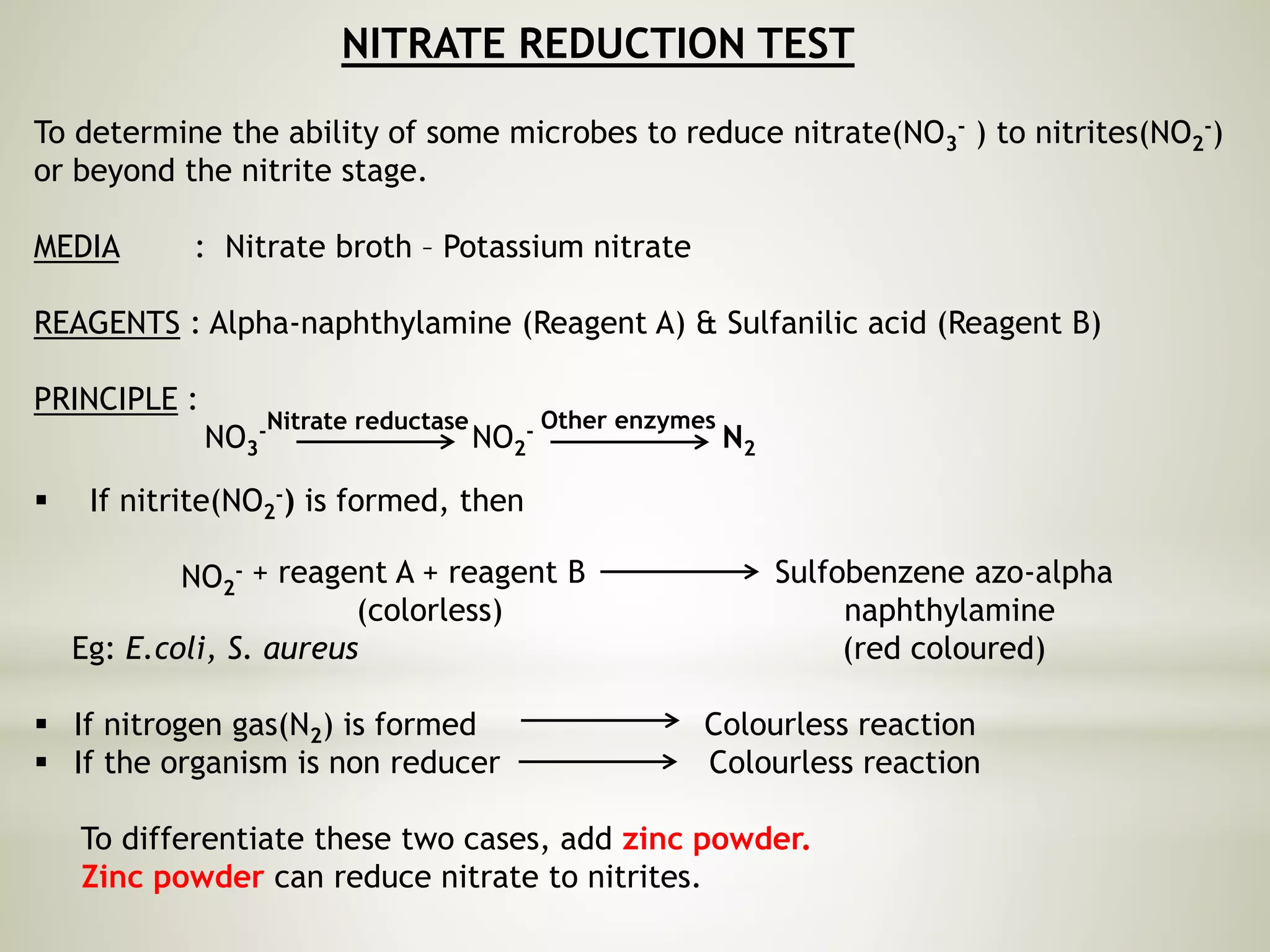
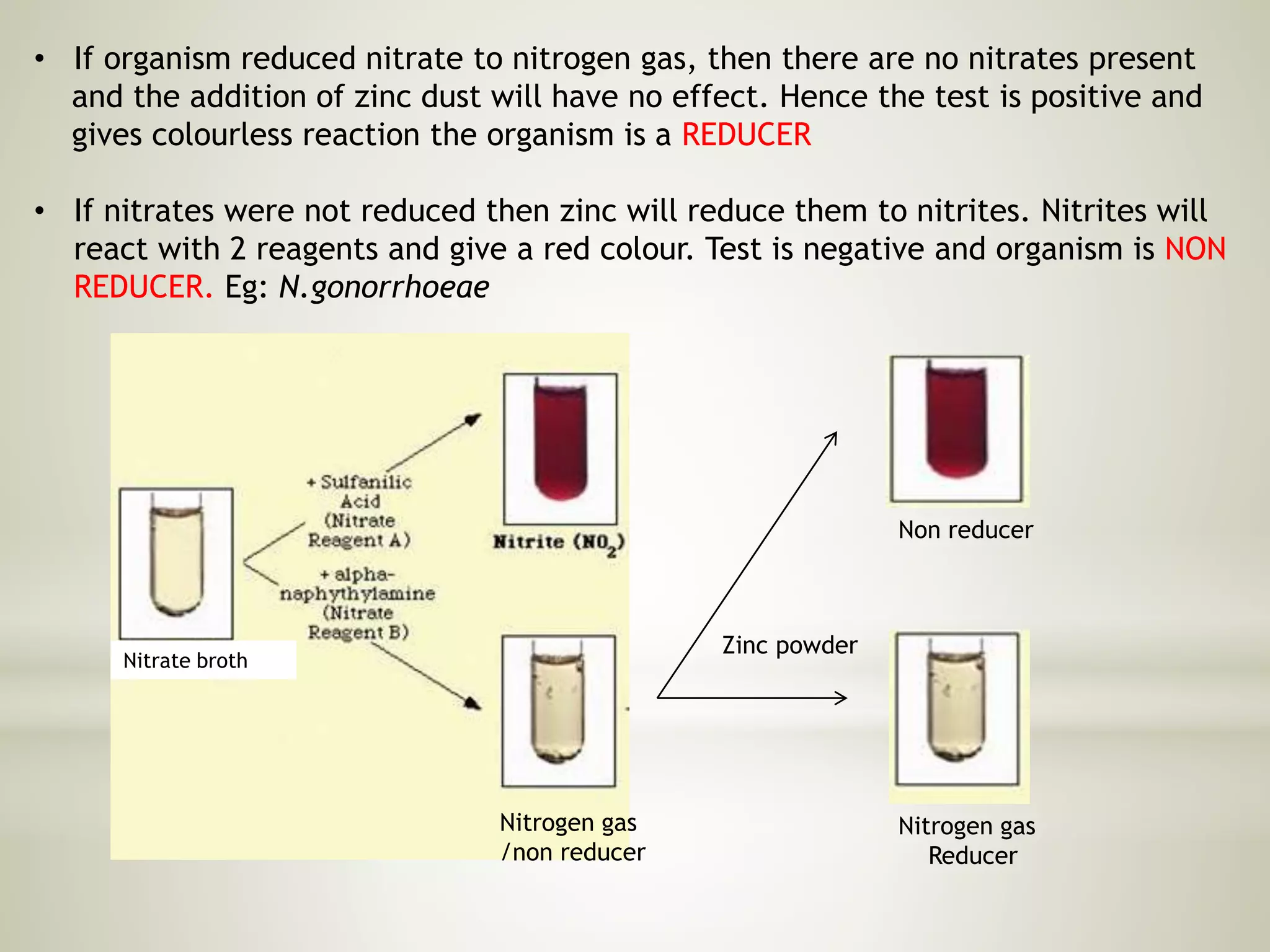
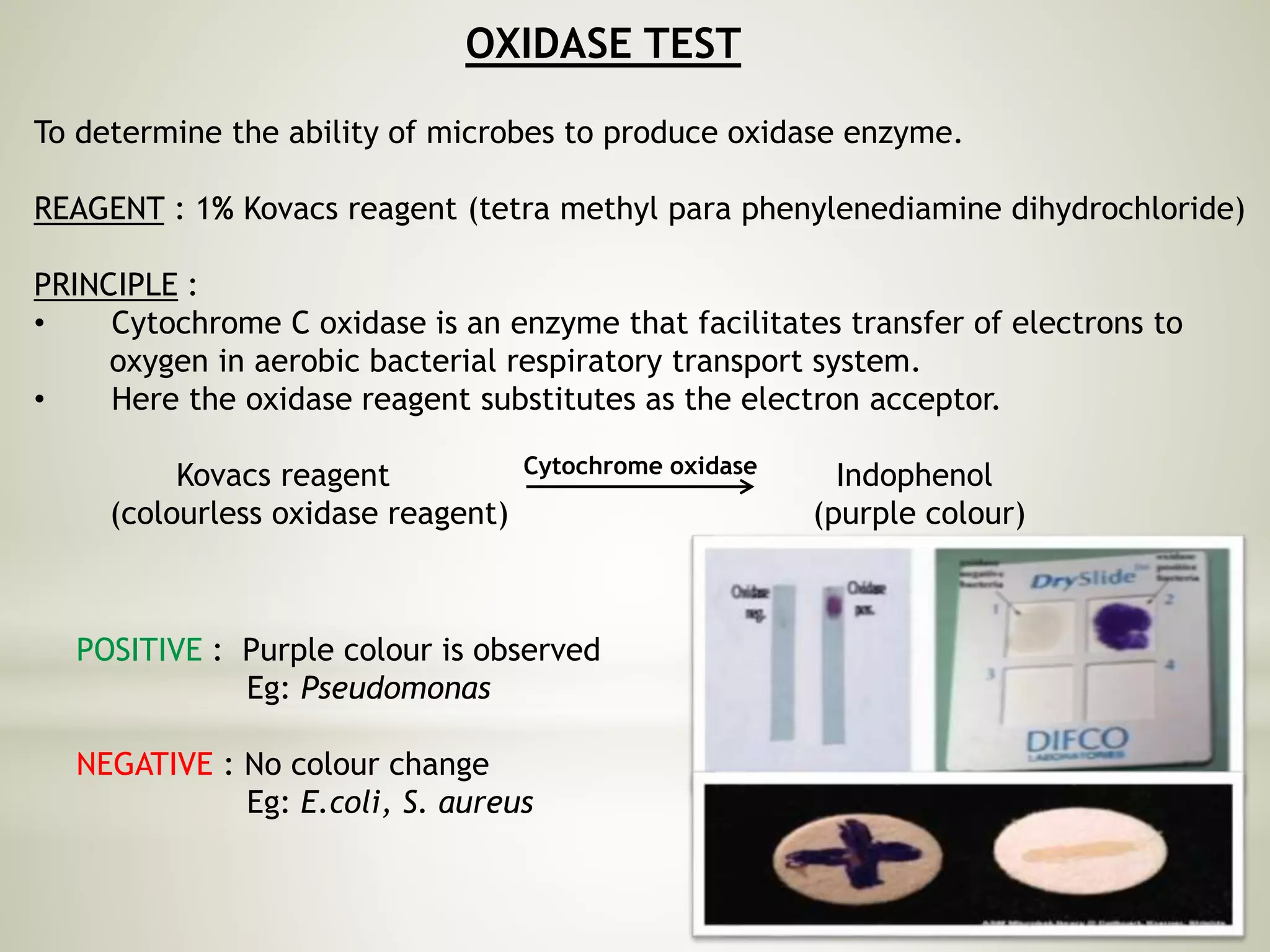
This document provides an overview of biochemical methods used for bacterial identification. It discusses why bacterial identification is important, the typical identification scheme involving isolation, staining, culturing, and biochemical/molecular tests. Several common biochemical tests are described in detail, including their principles, media used, and how to interpret results. These tests analyze bacterial metabolism of carbohydrates, proteins, lipids and other compounds. Automated identification systems that can rapidly identify bacteria based on biochemical profiles are also mentioned.