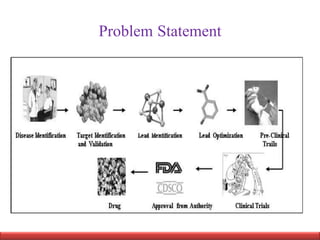
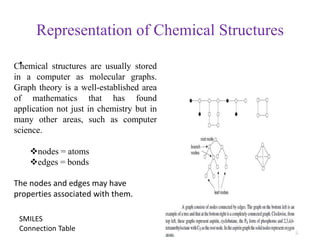
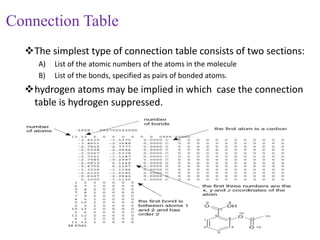
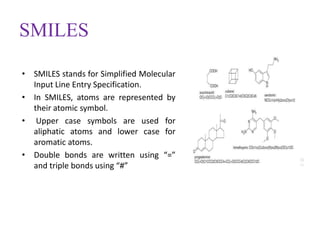
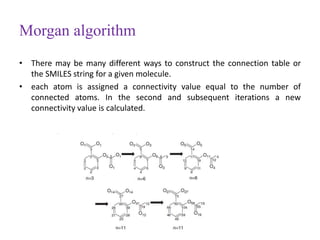
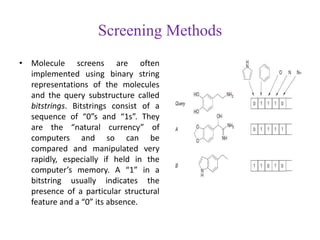
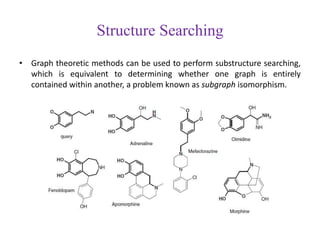
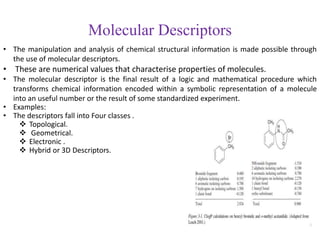
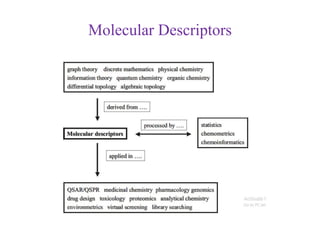
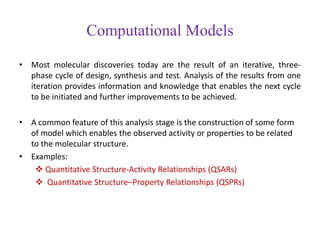
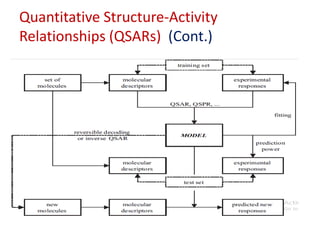
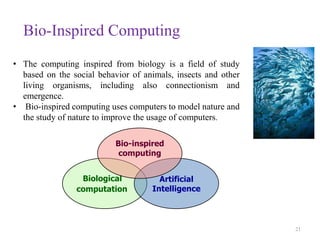
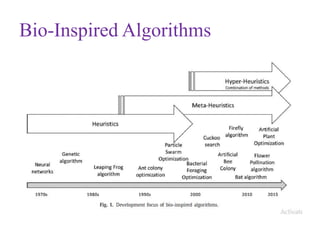
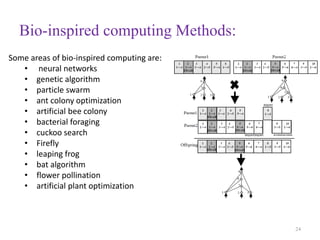
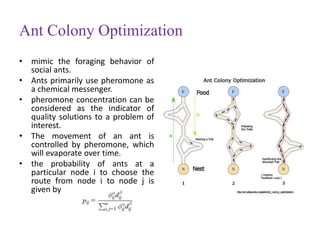
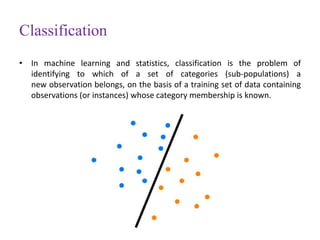
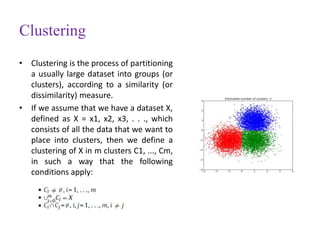
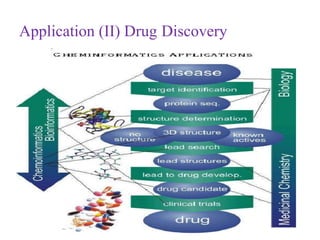
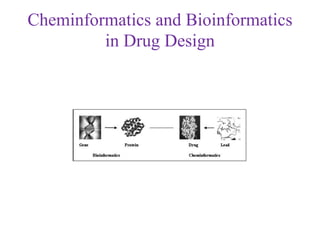
This document discusses applying bio-inspired computing techniques to problems in cheminformatics. It begins with introductions to cheminformatics and bio-inspired computing. Popular bio-inspired algorithms like ant colony optimization are explained. The document outlines applications of bio-inspired approaches to tasks in cheminformatics like classification, clustering, and feature selection. It concludes by noting potential applications in drug discovery and design.