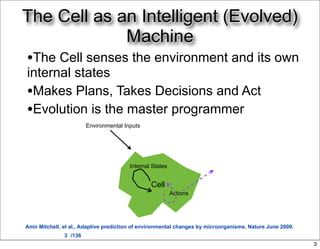
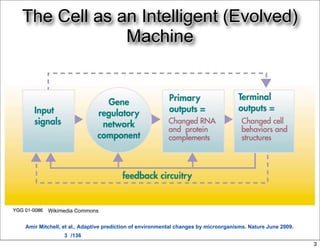
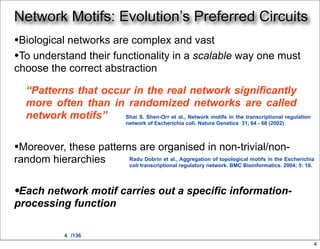
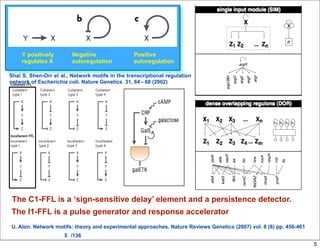
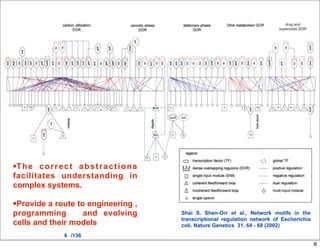
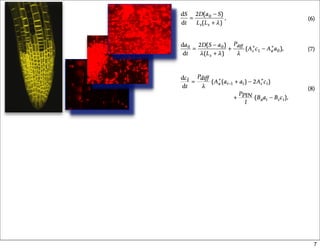
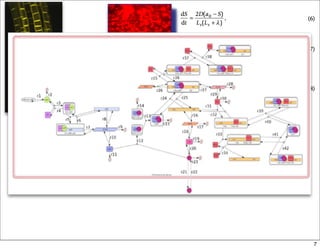
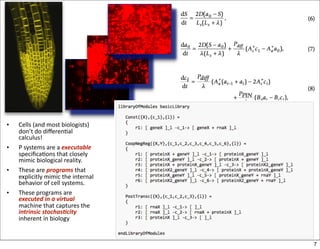
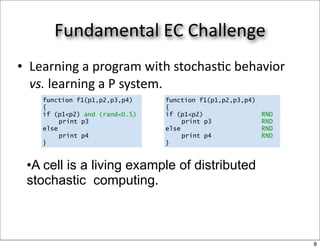
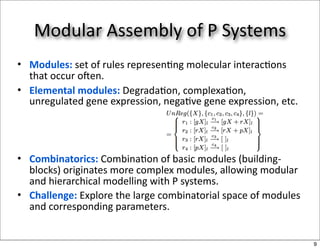
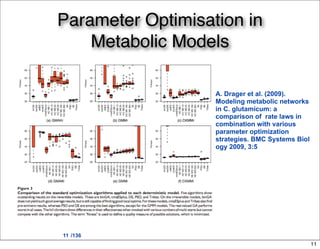
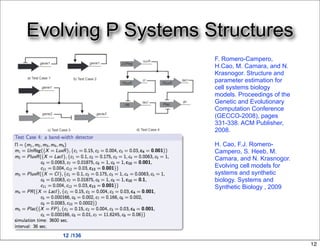
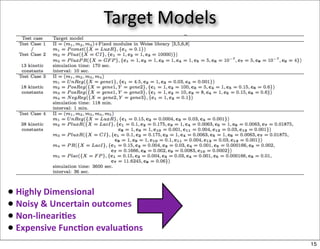
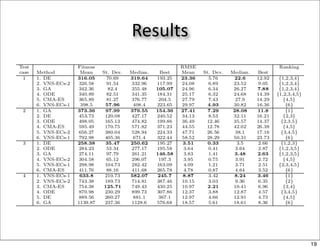
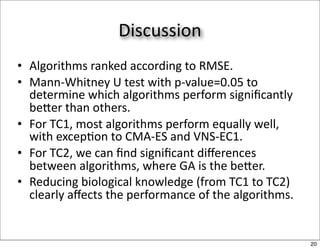
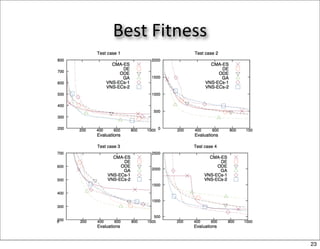
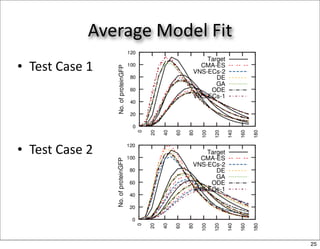
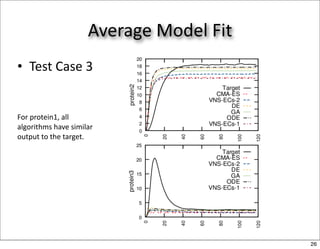
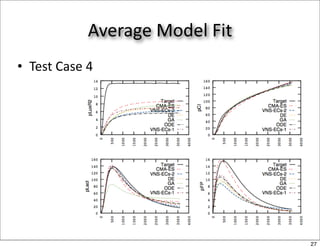
This document discusses using evolutionary algorithms to optimize parameters in P systems, which are computational models of biological cells. Four test cases of increasing difficulty are used to compare different algorithms. The results show that genetic algorithms, differential evolution, and opposition-based differential evolution perform better for problems with fewer parameters, while variable neighbourhood search algorithms perform better for the largest problem with 38 parameters. This is because the evolutionary algorithms are less efficient at optimizing large populations within the limited evaluation budget, whereas variable neighbourhood search focuses on a single solution.