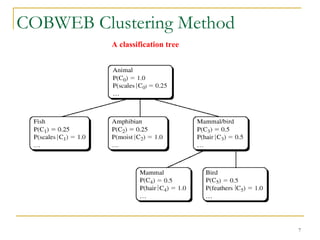
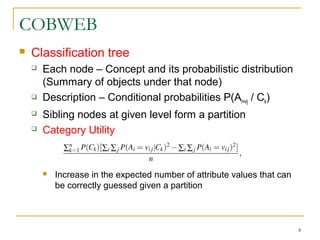
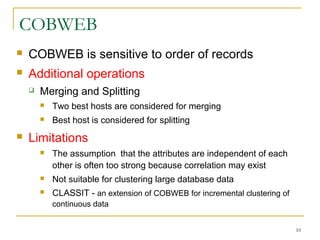
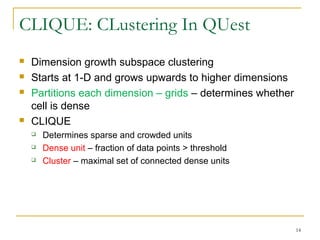
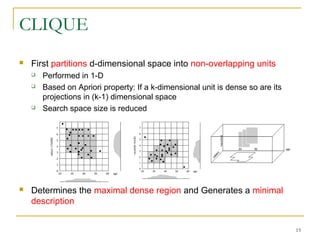
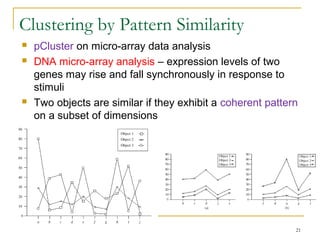
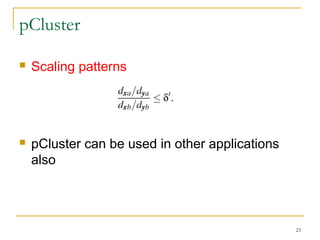
The document discusses various model-based clustering techniques for handling high-dimensional data, including expectation-maximization, conceptual clustering using COBWEB, self-organizing maps, subspace clustering with CLIQUE and PROCLUS, and frequent pattern-based clustering. It provides details on the methodology and assumptions of each technique.