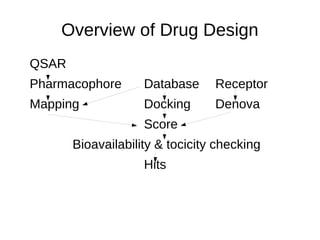
This document provides an overview of QSAR (Quantitative Structure-Activity Relationship) modeling, which establishes mathematical relationships between chemical structure descriptors of molecules and their biological activity. It discusses how QSAR analysis involves statistically analyzing properties of known active molecules to develop models that can then predict activity of new compounds. Examples of using QSAR to optimize ligand structures and select candidates for further testing are also provided.