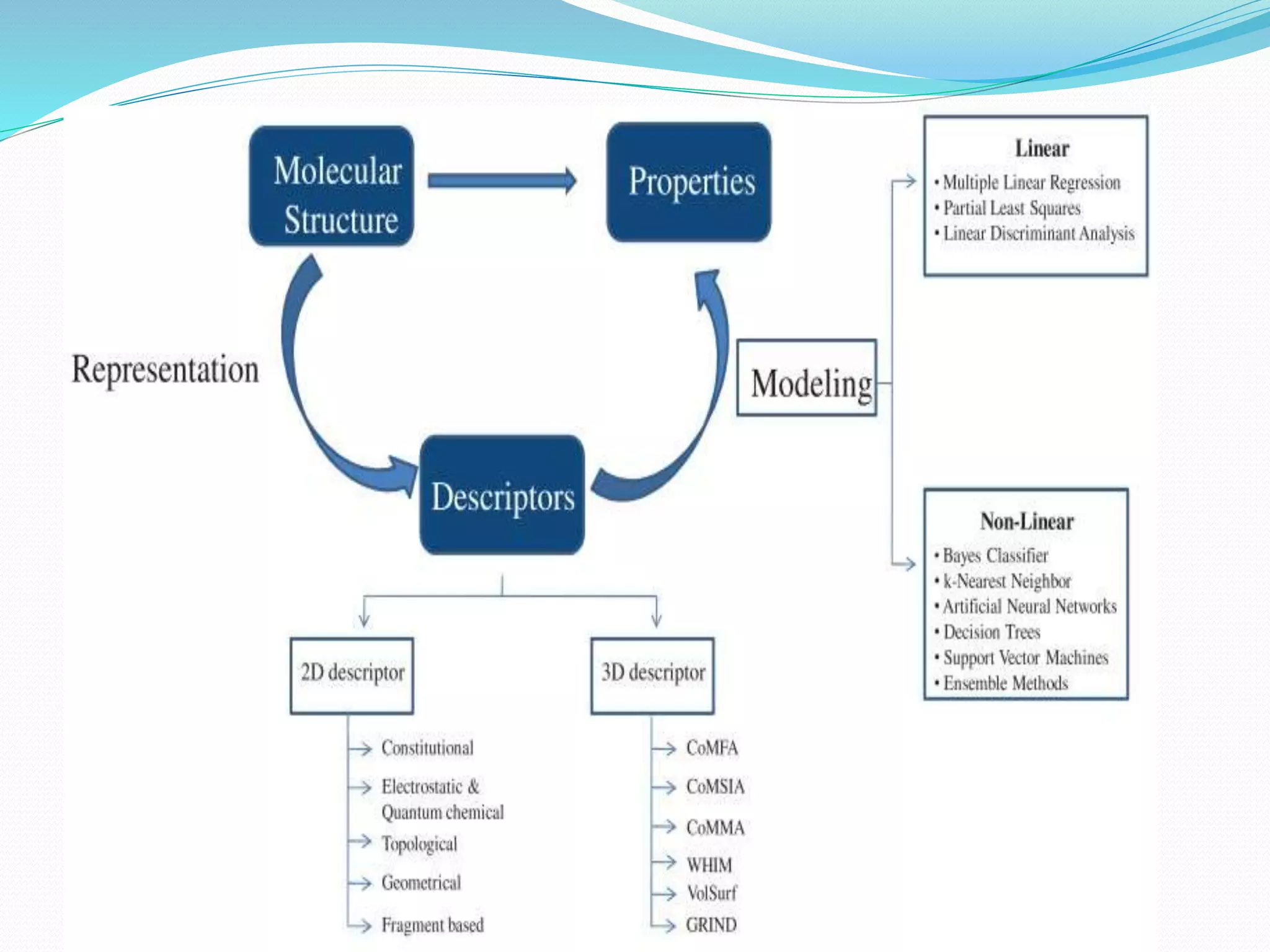
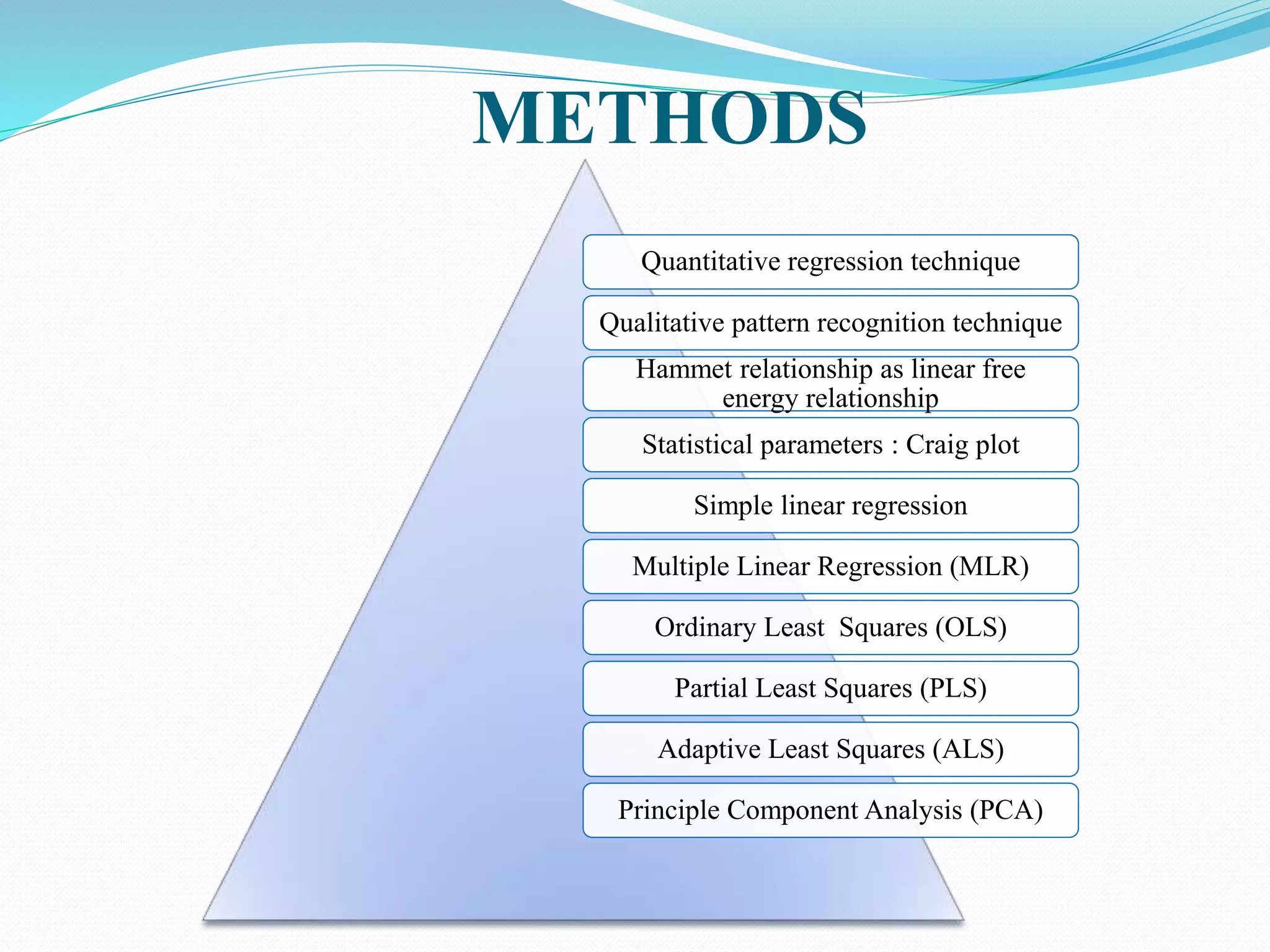
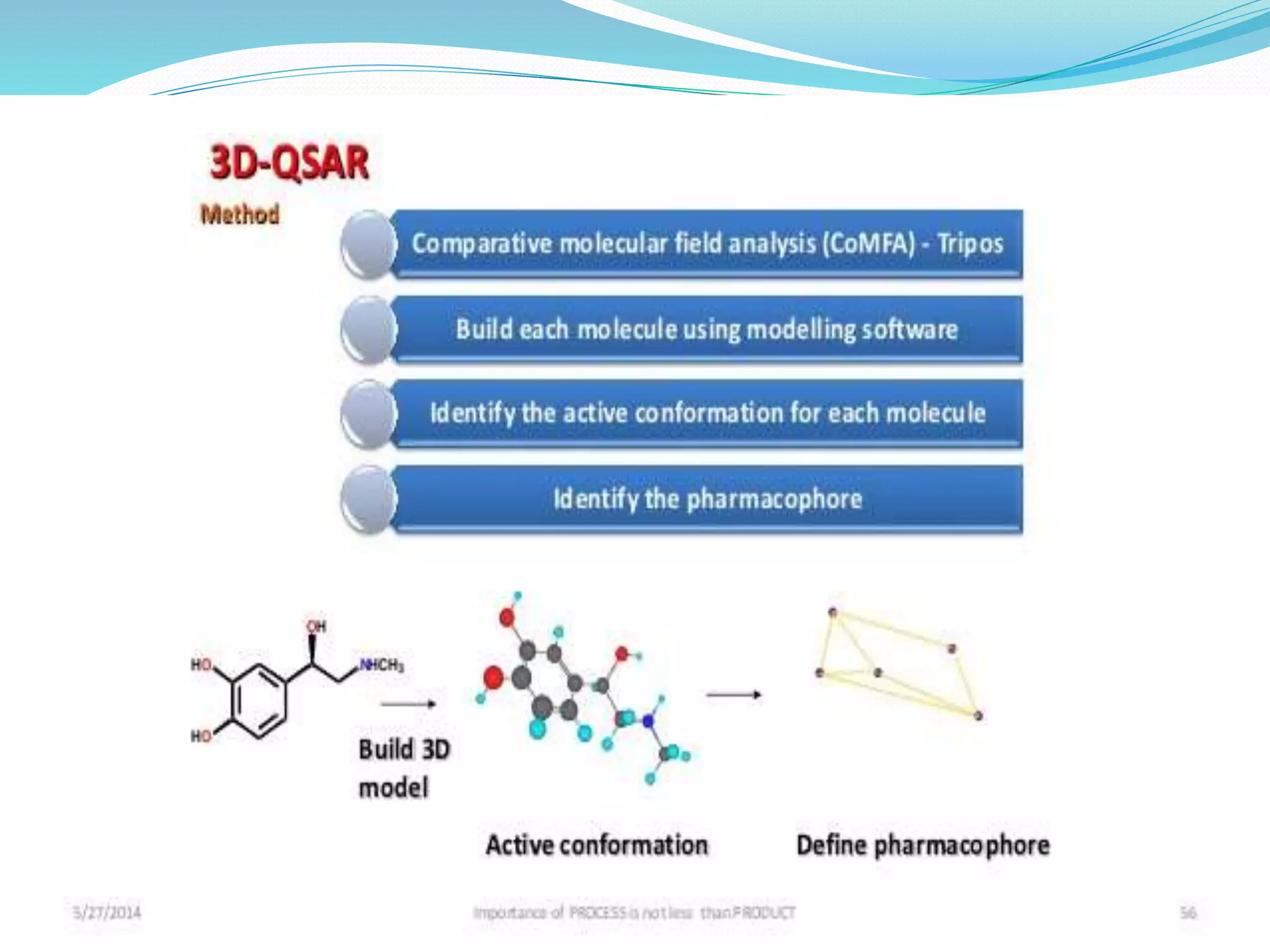
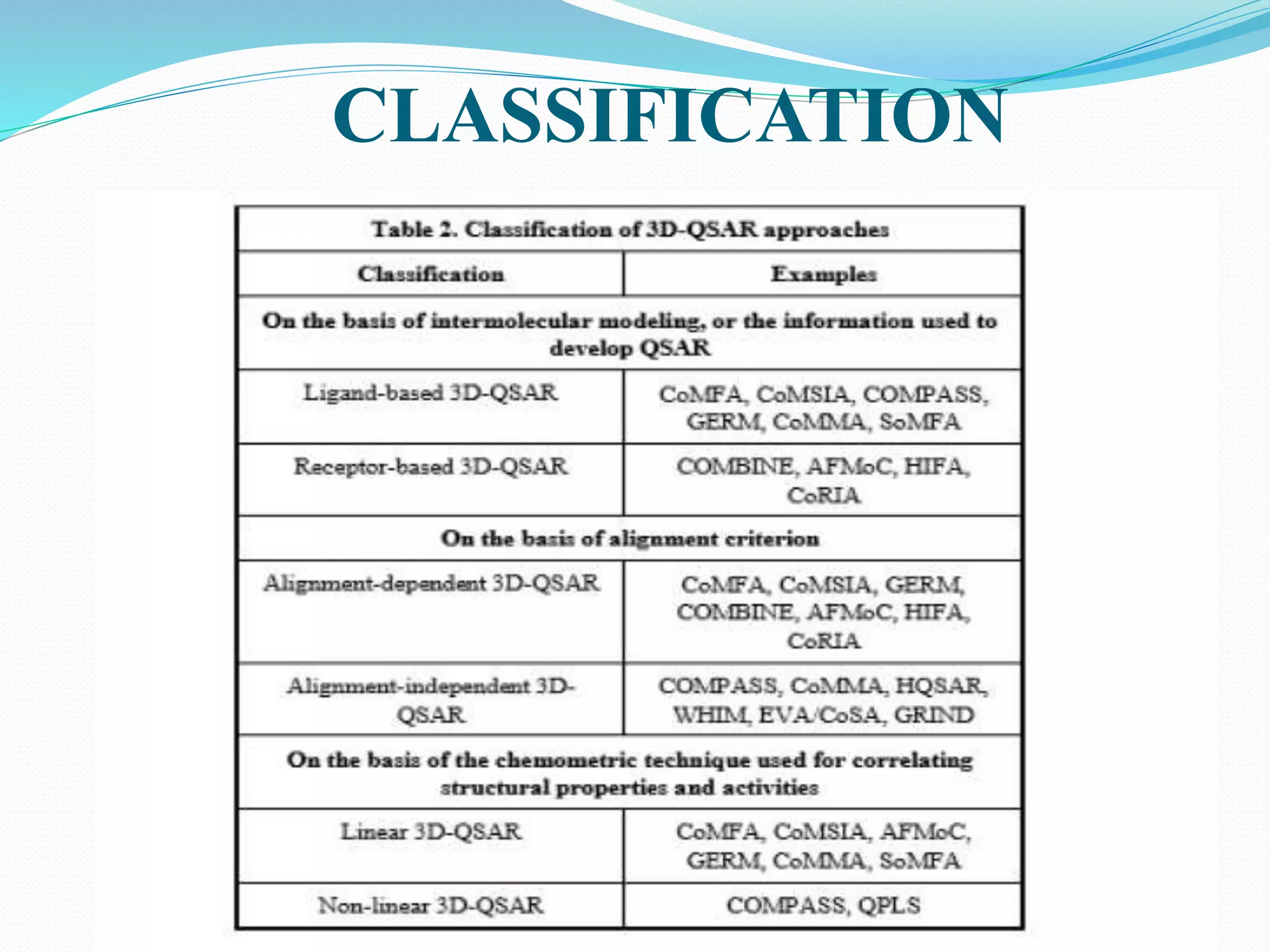
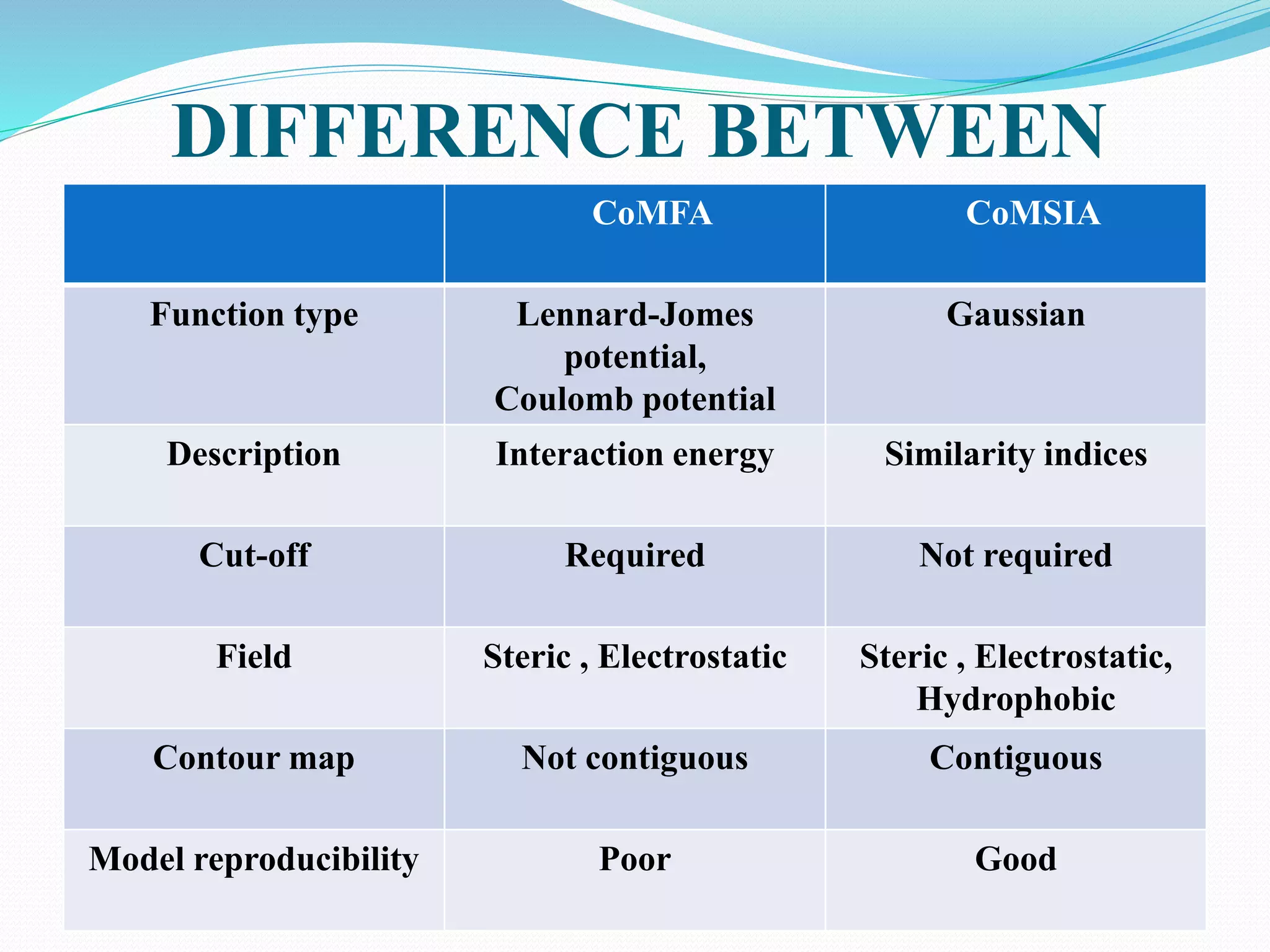
This document discusses quantitative structure-activity relationships (QSAR) modeling techniques. It introduces 2D-QSAR which uses molecular descriptors to correlate structure and activity. It also discusses 3D-QSAR techniques like CoMFA and CoMSIA which use 3D molecular fields/properties and statistical methods like PLS to model activity. These techniques are useful for drug design, virtual screening, and predicting absorption, distribution, metabolism, excretion properties.