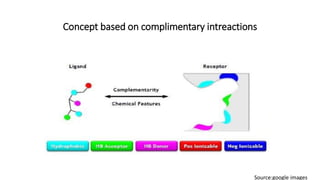
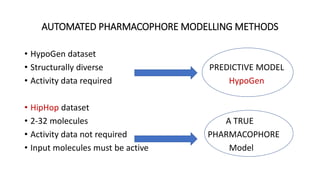
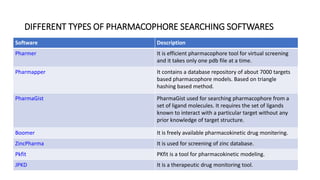
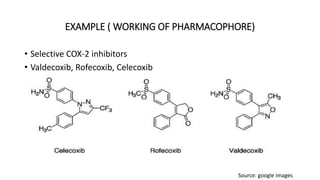
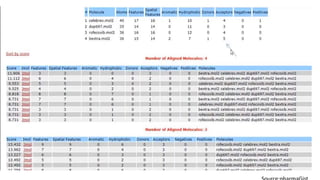
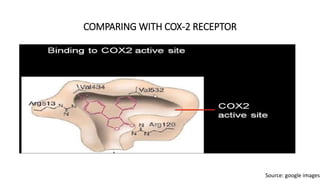
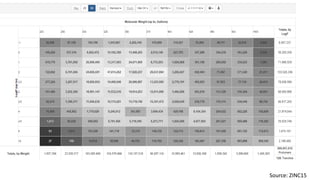
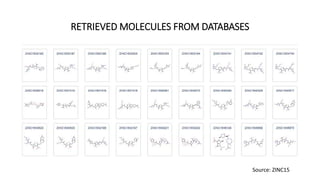
Pharmacophore modeling is an important technique in drug discovery. A pharmacophore model explains how structurally diverse ligands can bind to a common receptor site by identifying the essential chemical features necessary for biological activity. Pharmacophore models can be used in virtual screening to identify potential new drug leads from chemical databases that share the same features and binding orientation. Key steps in developing a pharmacophore model include selecting a training set of ligands, conformational analysis, molecular superimposition, and validation against biological activity data.