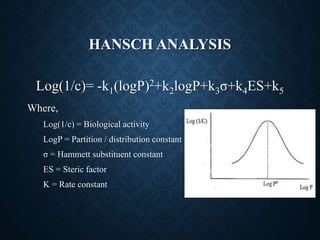
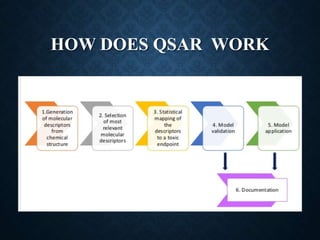
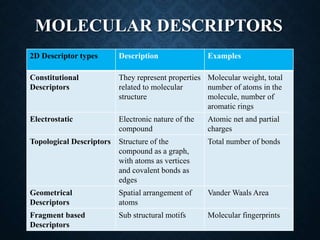
The document discusses 2D-QSAR (Quantitative Structure-Activity Relationship) analysis methods. It defines QSAR as mathematical relationships linking chemical structure and pharmacological activity. It describes several common 2D-QSAR methods including Hansch analysis, Free Wilson analysis, and various statistical methods. Cluster analysis is discussed as a way to group similar molecules and select a diverse subset for analysis. Molecular descriptors that encode structural, electronic, and topological properties are also introduced.