This document provides information about biological databases including:
- Different types of biological databases such as relational, object-oriented, hierarchical, and hybrid systems.
- Common uses of biological databases including annotation searches, homology searches, pattern searches, predictions, and comparisons.
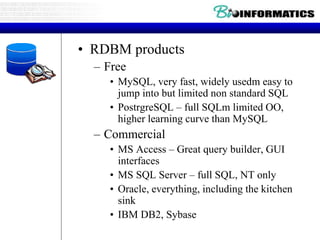
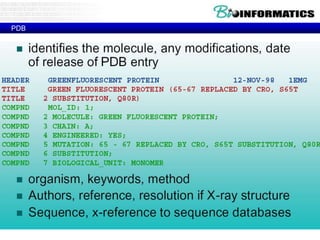
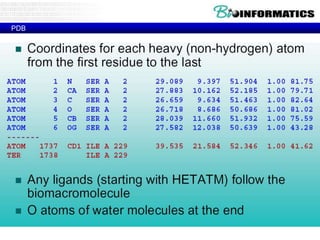
- Examples of database entries in common formats like GenBank, EMBL, and SwissProt that show the layout and key fields.


















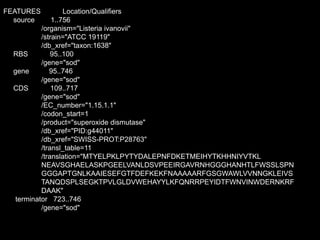


![EMBL format
ID LISOD standard; DNA; PRO; 756 BP. IDentification
XX
AC X64011; S78972; Accession (Axxxxx, Afxxxxxx), GUID
XX
NI g44010 Nucleotide Identifier --> x.x
XX
DT 28-APR-1992 (Rel. 31, Created) DaTe
DT 30-JUN-1993 (Rel. 36, Last updated, Version 6)
XX
DE L.ivanovii sod gene for superoxide dismutase DEscription
XX.
KW sod gene; superoxide dismutase. KeyWord
XX
OS Listeria ivanovii Organism Species
OC Eubacteria; Firmicutes; Low G+C gram-positive bacteria; Bacillaceae;
OC Listeria. Organism Classification
XX
RN [1]
RA Haas A., Goebel W.; Reference
RT "Cloning of a superoxide dismutase gene from Listeria ivanovii by
RT functional complementation in Escherichia coli and
RT characterization of the gene product.";
RL Mol. Gen. Genet. 231:313-322(1992).
XX](https://image.slidesharecdn.com/20160223biologicaldatabasespart1-160222224850/85/2016-02-23_biological_databases_part1-32-320.jpg)
![Example of a SwissProt entry
ID TNFA_HUMAN STANDARD; PRT; 233 AA. IDentification
AC P01375; ACcession
DT 21-JUL-1986 (REL. 01, CREATED) DaTe
DT 21-JUL-1986 (REL. 01, LAST SEQUENCE UPDATE)
DT 15-JUL-1998 (REL. 36, LAST ANNOTATION UPDATE)
DE TUMOR NECROSIS FACTOR PRECURSOR (TNF-ALPHA) (CACHECTIN).
GN TNFA. Gene name
OS HOMO SAPIENS (HUMAN). Organism Species
OC EUKARYOTA; METAZOA; CHORDATA; VERTEBRATA; TETRAPODA; MAMMALIA;
OC EUTHERIA; PRIMATES. Organism Classification
RN [1] Reference
RP SEQUENCE FROM N.A.
RX MEDLINE; 87217060.
RA NEDOSPASOV S.A., SHAKHOV A.N., TURETSKAYA R.L., METT V.A.,
RA AZIZOV M.M., GEORGIEV G.P., KOROBKO V.G., DOBRYNIN V.N.,
RA FILIPPOV S.A., BYSTROV N.S., BOLDYREVA E.F., CHUVPILO S.A.,
RA CHUMAKOV A.M., SHINGAROVA L.N., OVCHINNIKOV Y.A.;
RL COLD SPRING HARB. SYMP. QUANT. BIOL. 51:611-624(1986).
RN [2]
RP SEQUENCE FROM N.A.
RX MEDLINE; 85086244.
RA PENNICA D., NEDWIN G.E., HAYFLICK J.S., SEEBURG P.H., DERYNCK R.,
RA PALLADINO M.A., KOHR W.J., AGGARWAL B.B., GOEDDEL D.V.;
RL NATURE 312:724-729(1984).
...](https://image.slidesharecdn.com/20160223biologicaldatabasespart1-160222224850/85/2016-02-23_biological_databases_part1-33-320.jpg)









![Relational Database Terminology
Relational operators
• Relational
– select
rel WHERE boolean-xpr
– project
rel [ attr-specs ]
– join
rel JOIN rel
– divide by
rel DIVIDEBY rel
• Set-based
rel UNION rel
rel INTERSECT rel
rel MINUS rel
rel TIMES rel](https://image.slidesharecdn.com/20160223biologicaldatabasespart1-160222224850/85/2016-02-23_biological_databases_part1-53-320.jpg)